Single-cell metabolic imaging and digital scoring of fat tissue remodeling by label-free metabolic imaging
Single-cell metabolic imaging and digital scoring of fat tissue remodeling by label-free metabolic imaging
Kim, M.; Berger, C.; Wolf, A.; Peteranderl, A.; Klingenspor, M.; Ntziachristos, V.; Li, Y.; Pleitez, M. A.
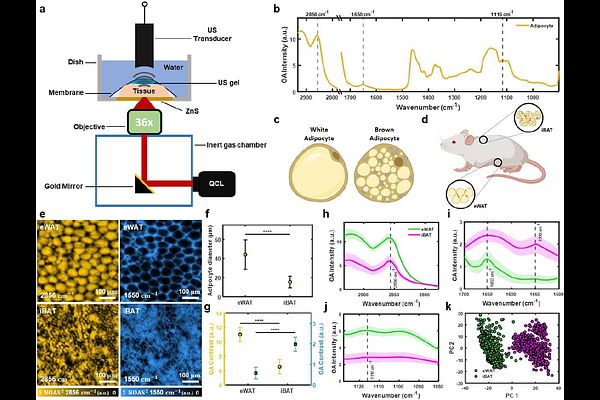
AbstractAdipose tissue plasticity and functional heterogeneity play a central role in maintaining energy homeostasis, and their malfunction leads to metabolic disorders such as obesity, diabetes, and cardiometabolic disease. Rapid, single-cell metabolic imaging of intact fat tissue not only extends our understanding of metabolic dynamics and heterogeneity but also holds great potential as a tool for clinical diagnosis. However, the use of exogenous labels and dyes in conventional optical microscopy results in tissue deformation and requires time-consuming tissue preparation. Here, we demonstrated single-cell imaging of metabolic changes and heterogeneity in freshly excised adipose tissues that can distinguish tissue types without the need for exogenous labels using bond-specific, non-destructive, mid-infrared optoacoustic microscopy (MiROM) that allows preserving the native tissue architecture with minimal sample preparation time. Further leveraging MiROM, we monitored intracellular molecular and morphological changes during postnatal remodeling of adipose tissue when metabolic characteristics of adipocytes undergo a transient drastic change. Additionally, we developed an AI-based quantitative spatial tissue analysis tool (Q-SAT) to predict the spatial distribution of white fat- and brown fat-like features, providing a robust digital scoring method for adipose tissue phenotypic assessment. Collectively, we implemented MiROM as an enabling technology to provide fast, label-free metabolic imaging of unprocessed adipose tissue, opening a new perspective for understanding and characterizing the morpho-functional dynamics of adipose tissue remodeling.