Farmed oyster mortality follows consistent Vibrio community reorganization
Farmed oyster mortality follows consistent Vibrio community reorganization
Smith, S.; Ciesielski, M.; Clerkin, T.; Ben-Horin, T.; Noble, R. T.
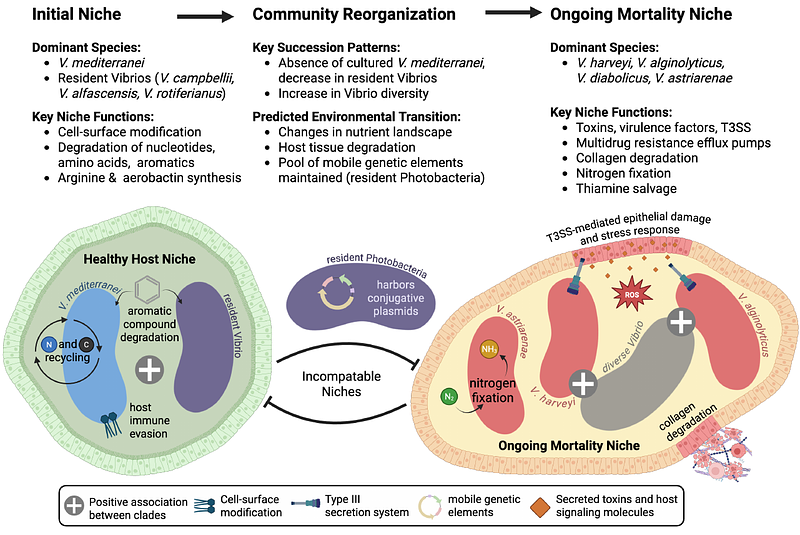
AbstractMortality events in marine bivalves cause substantial economic losses in aquaculture, yet the microbial dynamics underlying these events remain poorly characterized. Here, we investigated succession patterns in oyster-associated Vibrio communities during mortality events by sampling eastern oysters (Crassostrea virginica) at a North Carolina commercial farm that has experienced repeated, unexplained mortality events. Through whole-genome sequencing of 110 Vibrio isolates from 26 oysters sampled across mortality events in two consecutive years, we identified six conserved phylogenetic clades with distinct temporal associations. Vibrio mediterranei and a clade of resident Vibrios consistently dominated the initial cultured community at the onset of mortality. However, V. mediterranei was absent as mortality progressed, coinciding with increased abundance of V. harveyi, V. alginolyticus, V. diabolicus, and V. astriarenae. Comparative genomic analysis revealed that initial isolates were enriched in pathways associated with host colonization and complex carbon metabolism, while isolates from elevated mortality showed enrichment in virulence mechanisms and adaptation to degraded host tissues. Temporal separation between genetically distinct clades suggests microbial competition shapes community assembly during mortality events that ultimately reached >85% in both years. This predictable succession from commensal to potentially pathogenic Vibrio species provides genome-level insight into microbial community dynamics during oyster mortality. The consistent absence of V. mediterranei prior to severe mortality suggests this species could serve as a bioindicator for early warning systems to mitigate economic losses in shellfish aquaculture.