Macroevolutionary Rates of Species Interactions: Approximate Bayesian Inference from Cophylogenies
Macroevolutionary Rates of Species Interactions: Approximate Bayesian Inference from Cophylogenies
Zeng, Y.; Roman-Palacios, C.
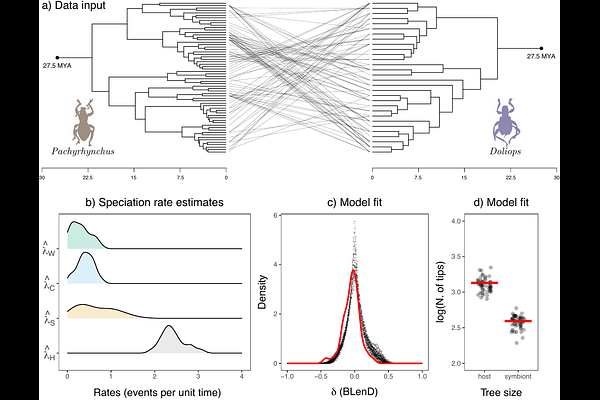
AbstractUnderstanding the macroevolutionary dynamics of species interactions such as parasitisms, commensalisms, and mutualisms is an important goal in evolutionary ecology. To this end, statistical inference from extant cophylogenetic systems holds immense potential. However, such inference cannot yet handle the speciation-extinction dynamics that occur simultaneously in the host and symbiont clades on the same timescale. Here we present an Approximate Bayesian Computation (ABC) approach that, while taking into account host and symbiont extinction, infers rates of four types of speciation from a cophylogenetic system: (i) host speciation, (ii) symbiont speciation without host-switching, (iii) symbiont speciation with host-switching, and (iv) cospeciation. The new ABC approach relies on a novel design of summary statistics combining both size-based (i.e., tree sizes) and size-free summary statistics (i.e., the normalized distribution of Branch Length Differences - BLenD) of the cophylogeny. Convergence analyses show that the combined design of summary statistics outperforms size-based or size-free summary statistics alone - achieving satisfactory accuracy in detecting rate heterogeneity between the four types of speciation. Our ABC approach allows the user to infer the predominant mode of speciation within a given cophylogenetic system. The approach is demonstrated with an application to a cophylogenetic dataset of commensalism, in which beetles of one genus mimic those of another. In this system, we identify host speciation as the predominant process with the fastest rate (in events per unit time) among all four types of speciation (i.e., 4.3-5.1 times faster than the median among all four types of speciation). Understanding how and why different types of speciation may predominate in different cophylogenetic systems can have implications for various areas in ecology and evolution such as host conservatism, trait-driven diversification, pathogen spillover risk, and parasite extinction risk. This new approach highlights the need and potential for future efforts to compile time-calibrated cophylogenies.