Multiparent Recombinant Inbred lines crossed to a tester provide novel insights into sources of cis and trans regulation of gene expression
Multiparent Recombinant Inbred lines crossed to a tester provide novel insights into sources of cis and trans regulation of gene expression
Marroni, F.; Morse, A. M.; Nanni, A.; Nolte, N. F.; Williams-Simon, P.; Leon-Novelo, L. G.; Graze, R. M.; Schmidt, P.; King, E. G.; McIntyre, L. M.
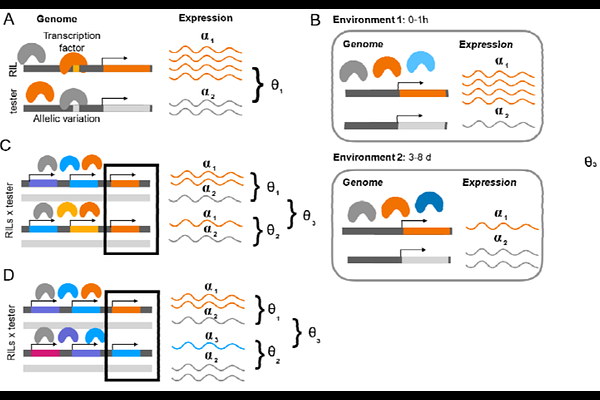
AbstractWe propose crossing multi-parent recombinant-inbred-lines (RILs) to a common tester and measuring allele specific gene expression in the offspring. With a few RIL x Tester crosses, genetic variation in the regulation of gene expression between RIL alleles can be identified. Testing whether allelic imbalance between two RIL x Tester crosses is equal, is a test of cis or trans depending on the RIL alleles compared. We demonstrate this approach in a long-read RNA-seq experiment in female abdominal tissue at two time points in Drosophila melanogaster. Among the 40% of all loci that show evidence of genetic variation in cis, trans effects due to the environment are detectable in 31% of loci and trans effects due to genetic background are detectable in 19% of loci with little overlap in sources of trans variation. The loci identified in this study are associated with loci previously reported to exhibit genetic variation in gene expression in a range of tissues and large population samples, suggesting that there is standing variation for genetic regulation of gene expression suggesting a potential source of cryptic genetic variation by which pleiotropic effects respond to changes in the environment. For loci in a QTL for thermotolerance, previously shown to differ in expression based on temperature; eleven of these loci had evidence for regulation of gene expression regardless of the environment, including Cpr67B, a cuticular protein. This study provides a blueprint for efficiently identifying alleles in multi-parent populations that contribute to regulatory variation in gene expression.