Dietary Protein Sources Shape Gut Microbiome: Insights from Meta-analysis and Machine Learning
Dietary Protein Sources Shape Gut Microbiome: Insights from Meta-analysis and Machine Learning
ADEJUMO, S. A.; Das, P.; Ghimire, S.; Kin Yun Lim, C.; Malas, J.; Allen, J.; Hampton-Marcell, J. A.
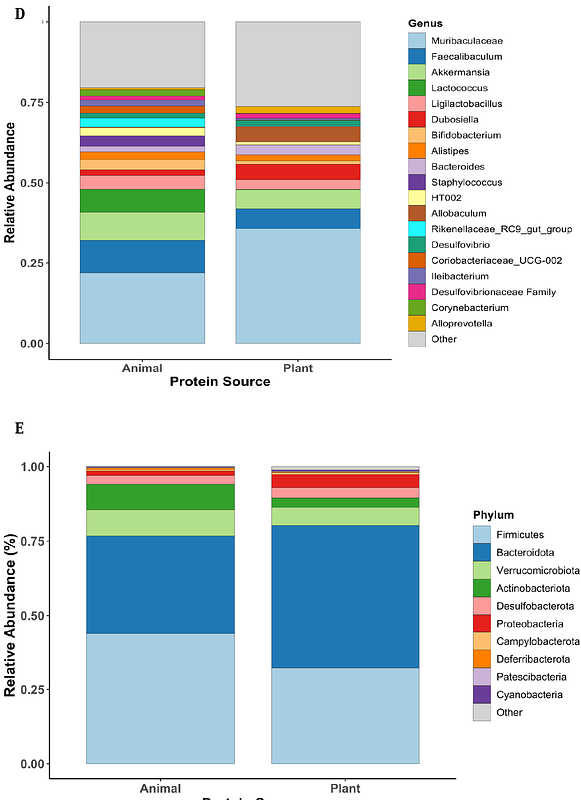
AbstractBackground: Dietary proteins, specifically protein fermentation, are commonly associated with gut microbiota composition; however, identifying microbial features that influence microbial diversity, composition, and structure and their implications in shaping host metabolism remains understudied and poorly understood. Plant- and animal-based proteins differ in amino acid composition and digestibility, yet their impact on microbial community structure and taxonomic shifts lacks systematic analysis. This meta-analysis applies machine learning to identify microbial taxa and their importance in predicting plant- versus animal-based protein diets in murine models, providing insights into bacterial contributions to protein metabolism and gut ecosystem stability. Methods: Amplicon sequencing data and metadata from 10 peer-reviewed murine microbiota studies (n = 187 mice) were analyzed to investigate the effects of plant- and animal-based protein diets. Study selection and data extraction followed PRISMA guidelines. 16S rRNA gene sequencing data were used to assess microbial community structure, diversity, and composition. Alpha diversity (Shannon, Simpson, Chao1) was analyzed using the Wilcoxon Rank-Sum test, while beta diversity was assessed using Aitchison distance with PCA for ordination. ANOSIM and PERMANOVA tested for statistical differences. Random Forest classification and LEfSe analysis identified microbial taxa significantly associated with dietary protein sources (p < 0.05, LDA > 3.0). Results: Plant-protein diets significantly increased microbial diversity (Shannon index, ANOVA, p < 0.001) and enriched 33 genera, including Bacteroides, Muribaculaceae, and Allobaculum. In contrast, animal-protein diets enriched seven genera, such as Colidextribacter, Clostridium_sensu_stricto_1, and Rikenellaceae_g (p < 0.01). Random Forest classification achieved 96.4% accuracy, identifying key microbial signatures distinguishing dietary groups. LEfSe analysis confirmed these differences, reinforcing distinct microbial adaptations to dietary protein sources. Conclusion: This study identifies diet-specific microbial biomarkers, demonstrating how dietary protein sources shape gut microbiota composition. These findings highlight the role of diet-microbiome interactions and suggest potential applications for microbiome-targeted nutritional strategies.