Acquisition and repeated alteration of (TTGGG)n telomeric repeats in Odonata (dragonflies and damselflies)
Acquisition and repeated alteration of (TTGGG)n telomeric repeats in Odonata (dragonflies and damselflies)
Gotoh, T.; Suzuki, H.; Moriyama, M.; Futahashi, R.; Osanai-Futahashi, M.
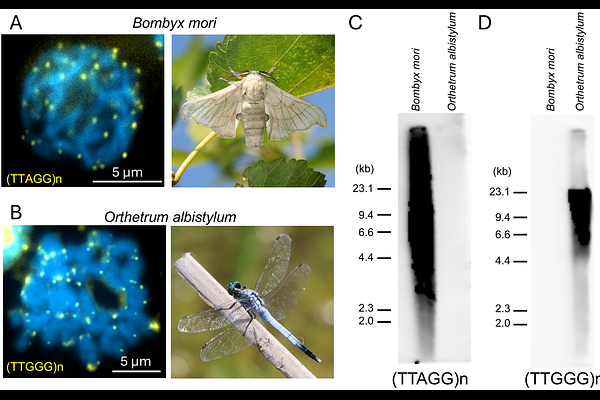
AbstractChromosome ends of most eukaryotes are composed of simple telomeric repeats. For arthropods, TTAGG pentanucleotide repeats, (TTAGG)n has been considered as the ancestral telomeric repeat. However, in the order Odonata, the earliest diverged group in insects that contains dragonflies and damselflies, (TTAGG)n signal has been almost undetectable in most examined species. Here, we report a unique pentanucleotide repeat (TTGGG)n as the typical telomeric repeat sequence for Odonata (dragonflies and damselflies). Based on genomic information from ten Odonata species, (TTGGG)n was considered the most likely candidate for telomeric repeat sequences. By fluorescence in situ hybridization (FISH) using 12 Odonata species, clear (TTGGG)n signals were detected at the chromosome ends in both dragonflies and damselflies. By Southern hybridization using 63 Odonata species, strong (TTGGG)n signals were detected from the majority of species covering all three suborders of Odonata, indicating that the telomeric repeat of the common ancestor of extant Odonata is (TTGGG)n. Notably, there were a few Odonata species in which (TTGGG)n signals were faint or absent, suggesting that the telomeric repeat sequence has repeatedly diverged in Odonata, even within genera such as Sympetrum. We also identified telomerase genes in both dragonflies and damselflies, and in some species, more than two telomerase genes are suggested to be present. Overall, this study demonstrates the ancestral acquisition of novel telomeric repeats and their repeated alteration in Odonata.