RamanMAE: Masked Autoencoders Enable Efficient Molecular Imaging by Learning Biologically Meaningful Spectral Representations
RamanMAE: Masked Autoencoders Enable Efficient Molecular Imaging by Learning Biologically Meaningful Spectral Representations
Paidi, S.; Maheshwari, P.
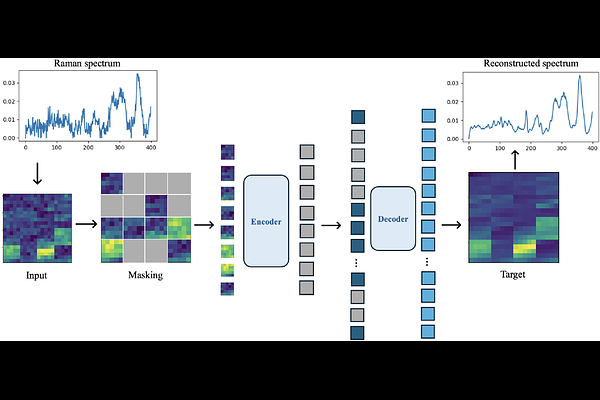
AbstractTraditional histopathological analysis of cells and tissue relies on morphological features from stained biopsy samples, which fail to leverage the wealth of chemical information about the underlying pathological states. Raman spectroscopy, a form of vibrational spectroscopy, uses light scattering to capture chemical information about the biological specimen. However, advancements in Raman spectroscopy are hindered by the method\'s intrinsic low throughput and the difficulty in deriving meaningful insights from the high-dimensional noisy datasets. In this paper, we propose RamanMAE, a spectral language model using masked autoencoders on large Raman spectral datasets from biological applications that can be used for spectral processing in applications with limited data. We achieved excellent reconstruction of masked patches of the spectra. We learned meaningful latent representations of the spectra that capture biological compositional information and serve as a low-dimensional feature space for building downstream machine learning methods. We showed that the decoder serves as an effective smoothing technique to reduce noise in the spectra and allow better localization and visualization of biological features in the spectral maps. We also demonstrated the transferability of the representations learned on one dataset to a different biological application.