SyntheMol-RL: a flexible reinforcement learning framework for designing novel and synthesizable antibiotics
SyntheMol-RL: a flexible reinforcement learning framework for designing novel and synthesizable antibiotics
Swanson, K.; Liu, G.; Catacutan, D. B.; McLellan, S.; Arnold, A.; Tu, M. M.; Brown, E. D.; Zou, J.; Stokes, J.
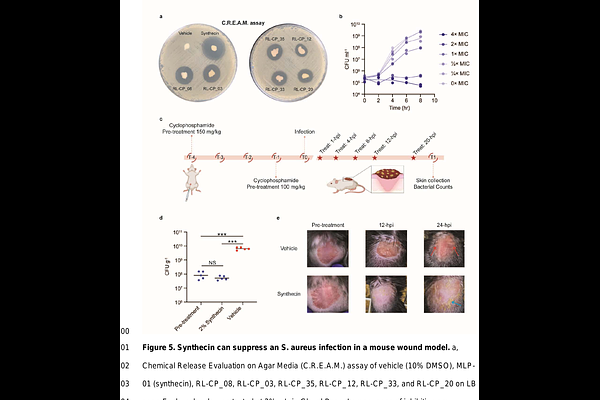
AbstractThe rise of antibiotic-resistant pathogens such as Staphylococcus aureus has created an urgent need for new antibiotics. Generative artificial intelligence (AI) has shown promise in drug discovery, but existing models often fail to propose compounds that are both effective and synthetically tractable. To address these challenges, we introduce SyntheMol-RL, a reinforcement learning-based generative model that can rapidly design synthetically accessible small molecule drug candidates from a massive chemical space of 46 billion compounds. SyntheMol-RL improves upon our prior Monte Carlo Tree Search (MCTS)-based SyntheMol model by generalizing across chemically similar building blocks and enabling multi-parameter optimization. We applied SyntheMol-RL to generate candidate antibiotics against S. aureus by optimizing for both antibacterial activity and aqueous solubility, and we found that SyntheMol-RL generated molecules with improved predicted properties compared to both the previous MCTS version of SyntheMol as well as an AI-based virtual screening baseline. We synthesized 79 SyntheMol-RL compounds that were unique relative to the training dataset and found that 13 showed potent in vitro activity, of which seven were structurally novel after detailed literature searches. Furthermore, one hit compound, synthecin, demonstrated efficacy in a murine wound infection model of methicillin-resistant S. aureus (MRSA). These results validate SyntheMol-RL\'s ability to generate novel and synthetically accessible candidate antibiotics and position SyntheMol-RL as a powerful tool for drug design across therapeutic domains.