Efficacy of natural marine sponges as a passive environmental DNA sampler for freshwater fish diversity monitoring
Efficacy of natural marine sponges as a passive environmental DNA sampler for freshwater fish diversity monitoring
Nakao, R.; Inaba, M.; Miyazono, S.; Saito, M.; Maruyama, K.; Imamura, F.; Akamatsu, Y.
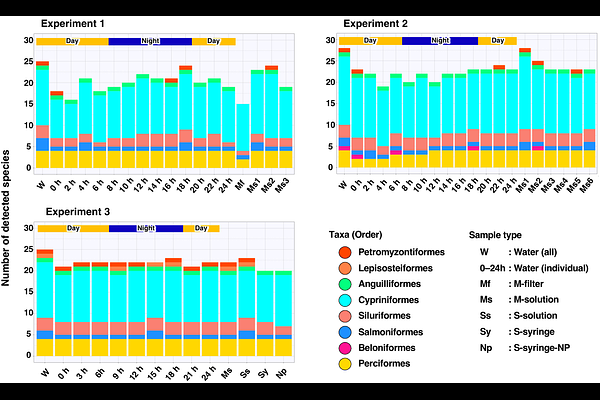
AbstractEnvironmental DNA (eDNA) analysis is a cost-effective and non-invasive tool for species and biodiversity monitoring in aquatic environments. Passive eDNA sampling method is focused as a novel and alternative to conventional sampling methods, such as water filtration. In this study, we examined the efficacy of the sponge skeleton as an eDNA passive sampler for the freshwater fish monitoring tool. We compared the performance of passive sampling method with standard water filtration in the river environment. A total of five different DNA extraction methods were also conducted in three experiments, and a suitable DNA extraction method from sponge skeleton was estimated. Quantitative fish metabarcoding using MiFish primers revealed no significant difference in species richness between water and passive sampling methods. While both sampling methods showed comparable trends in fish community structures, different clusters were identified for water sampling and passive samplers by the differences in DNA concentration of each fish species. Fish diversity in passive samples was comparable among the four DNA extraction methods using the Direct Capture method, except for the filtration method. Our results demonstrate the efficacy of passive eDNA sampling for monitoring freshwater fish diversity and potential use of sponge skeletons for the absorption material of the eDNA passive sampler.