ADAM-tRNA-seq: An Optimized Approach for Demultiplexing and Enhanced Hierarchal Mapping in Direct tRNA Sequencing
ADAM-tRNA-seq: An Optimized Approach for Demultiplexing and Enhanced Hierarchal Mapping in Direct tRNA Sequencing
Alarcon, R.; Koester, D.; Behrmann, S.; Ignatova, Z.
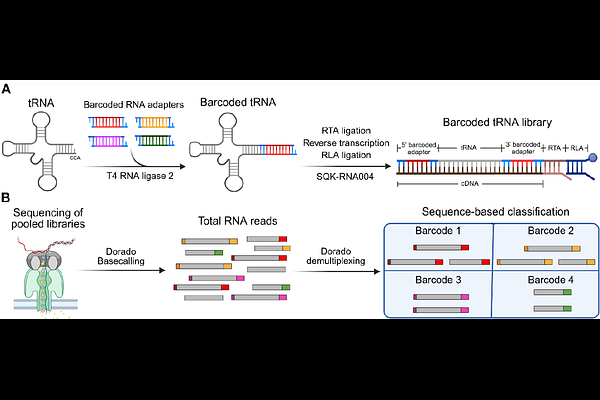
AbstractTransfer RNAs (tRNAs) play an essential role in protein synthesis and cellular homeostasis, with their dysregulation associated with various human pathologies. Recent advances in direct RNA sequencing by the Nanopore platform have enabled simultaneous profiling of tRNA abundance, modifications, and aminoacylation status. However, the high sequence similarity among tRNAs and the lack of robust demultiplexing strategies reduce the accuracy and limit the scalability of current approaches. Here, we developed ADAM-tRNA-seq, a framework that addresses two key limitations of the Nanopore-based direct tRNA sequencing. First, we develop an RNA-based barcode demultiplexing method, employing a barcode embedded within the sequencing adapter that is recognized by the Dorado basecaller. Second, we designed a hierarchy-based mapping strategy that mitigates read loss due to multimapping by classifying reads at the isodecoder, isoacceptor, or isotype levels, thereby enhancing quantification accuracy. We validated ADAM-tRNA-seq using both synthetic tRNAs and a complex human tRNA pool, and systematically optimized it to achieve up to 99% classification precision. Together, these developments enable more accurate, scalable, and comprehensive characterization of tRNA pools in diverse sample types.