Cellular and Immune Adaptations at the Maternal-Fetal Interface in Bats
Cellular and Immune Adaptations at the Maternal-Fetal Interface in Bats
Caldwell, A.; Yang, L.; Casazza, R. L.; Worota, R. E.; McCutcheon, C.; Creisher, P. S.; Zhan, E.; Reasoner, C.; Higgins, A.; Schountz, T.; Coyne, C. B.
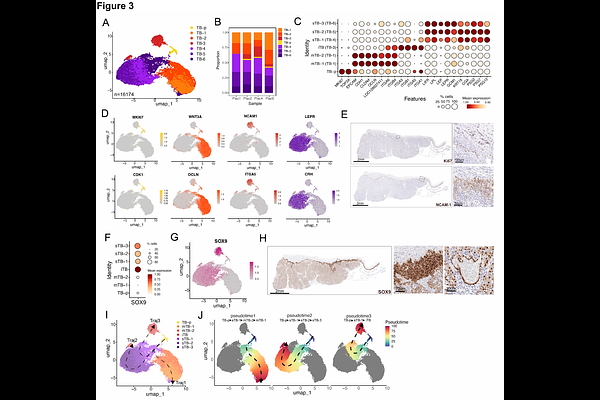
AbstractBats maintain pregnancy despite extended gestation relative to other small mammals, high fetal investment, recurrent pathogen exposure, and the metabolic demands of flight. These physiological extremes likely drive unique adaptations in placental function and maternal-fetal immune regulation, yet the cellular and molecular basis of these adaptations remains largely unknown. Here, we mapped the cellular landscape of the Jamaican fruit bat (Artibeus jamaicensis) placenta using single-nucleus RNA sequencing (snRNA-seq), integrated with histological and immunohistochemical analyses. We identified diverse trophoblast, stromal, and immune populations at the maternal-fetal interface, including specialized macrophages expressing pregnancy-associated signaling molecules. Trajectory analysis revealed dynamic trophoblast differentiation through proliferative, invasive, and syncytial states. To model these processes in vitro, we derived trophoblast and decidual gland organoids from matched tissues, which recapitulated key in vivo cell types and lineage trajectories. Cross-species transcriptomic comparisons with human and mouse placentas uncovered bat-specific gene programs in trophoblasts, fibroblasts, and immune cells. Notably, bat trophoblast organoids exhibited attenuated antiviral signaling compared to their human counterparts, suggesting species-specific modulation of innate immunity at the maternal-fetal interface. These findings define cellular strategies that support pregnancy under extreme physiological conditions and establish a framework for investigating the evolution of placental adaptations across mammals.