Eco-evolutionary feedbacks drive the co-occurrence of restriction-modification systems and antimicrobial resistance genes
Eco-evolutionary feedbacks drive the co-occurrence of restriction-modification systems and antimicrobial resistance genes
Westley, J.; Bedekar, P.; Pursey, E.; Szczelkun, M. D.; Recker, M.; van Houte, S.; Westra, E.
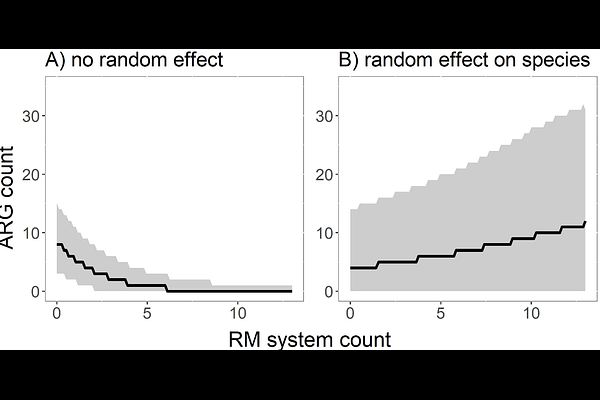
AbstractBacterial pathogens commonly become drug resistant via horizontal acquisition of antimicrobial resistance genes (ARGs), which are often encoded on mobile genetic elements (MGEs). Although bacterial defence systems are typically considered barriers to horizontal gene transfer (HGT), previous studies revealed that bacteria with more restriction-modification (RM) systems (the most abundant bacterial defences) frequently carry more MGEs. It was suggested that this counterintuitive relationship might result from stronger selection for RM systems when exposure to costly MGEs increases. Here, we test this hypothesis using a combination of modelling and bioinformatics analysis of >40,000 bacterial genomes to better understand how eco-evolutionary feedbacks between selection for RM and acquisition of MGEs shape bacterial genome evolution. Our model predicts negative associations between HGT and RM, but only if RM diversity is high. By contrast, at low RM diversity, eco-evolutionary feedbacks drive the emergence of positive associations between HGT and RM. Consistent with these predictions, we identified negative relationships between acquired ARG counts and RM counts across species but positive relationships within individual species. Collectively, our work helps to understand how RM systems shape patterns of HGT of ARGs, which may offer opportunities for targeted surveillance of strains at higher risk of horizontally acquiring novel drug resistance alleles.