Automated Registration and Clustering for Enhanced Localization Atomic Force Microscopy of Flexible Membrane Proteins
Automated Registration and Clustering for Enhanced Localization Atomic Force Microscopy of Flexible Membrane Proteins
Lisowski, C. M.; King, G. M.; Kosztin, I.
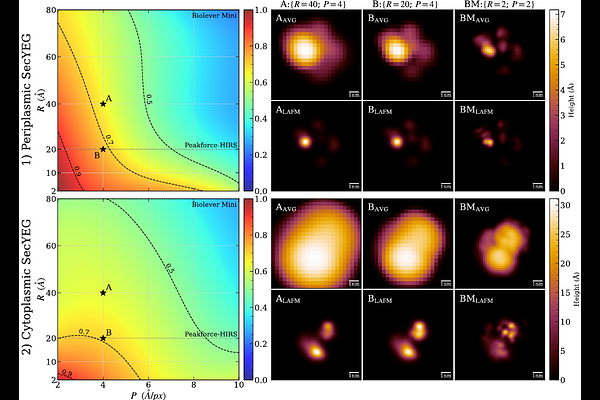
AbstractAtomic Force Microscopy (AFM) can create images of biomolecules under near-native conditions but suffers from limited lateral resolution due to the finite AFM tip size and recording frequency. The recently developed Localization Atomic Force Microscopy or LAFM (Heath et al., Nature 594, 385 (2021)) enhances lateral resolution by reconstructing peak positions in AFM image stacks, but it is less effective for flexible proteins with multiple conformations. Here we introduce an unsupervised deep learning algorithm that simultaneously registers and clusters images by protein conformation, thus making LAFM applicable to more flexible proteins. Using simulated AFM images from molecular dynamics simulations of the SecYEG translocon as a model membrane protein system, we demonstrate improved resolution for individual protein conformations. This work represents a step towards a more general LAFM algorithm that can handle biological macromolecules with multiple distinct conformational states such as SecYEG.