Stronger population suppression by gene drive targeting doublesex from dominant female-sterile resistance alleles
Stronger population suppression by gene drive targeting doublesex from dominant female-sterile resistance alleles
Chen, W.; Wang, Z.; Champer, J.
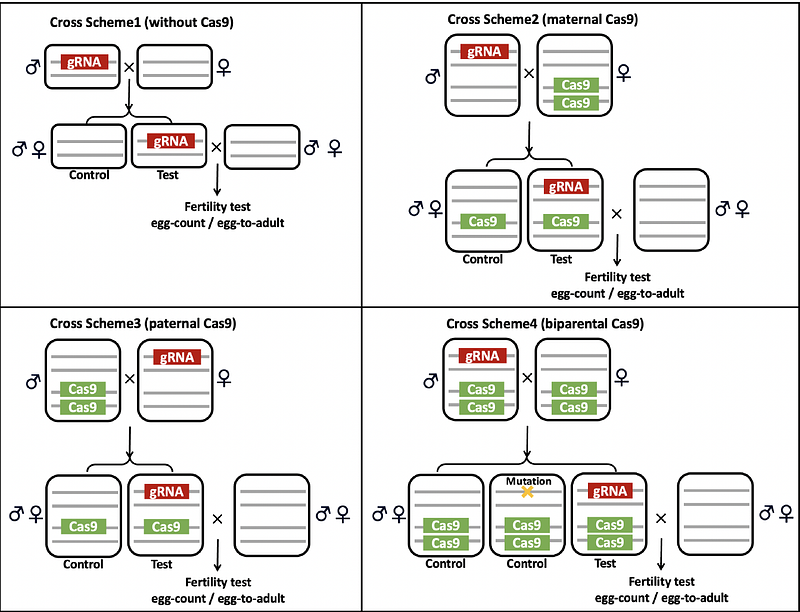
AbstractCRISPR homing drives can be used to suppress a population by targeting female fertility genes. They convert wild-type alleles to drive alleles in the germline of drive heterozygotes by homology-directed repair after DNA cleavage. However, resistance alleles produced by end-joining pose a great threat to homing drive. They prevent further recognition by Cas9, and therefore weaken suppressive power, or even stop suppression if they preserve the function of the target gene. We used multiplexed gRNAs targeting doublesex in Drosophila to avoid functional resistance and create resistance alleles that were dominant female-sterile. This occurred because the male dsx transcript was generated in females by disruption of the female-specific splicing acceptor site. We rescued dominant sterility of the drive by providing an alternate splicing site. As desired, the drive was recessive female sterile and yielded high drive inheritance among the progeny of both male and female drive heterozygotes. The dominant-sterile resistance alleles enabled stronger suppression in computational models, even in the face of modest drive efficiency and fitness costs. However, we found that male drive homozygotes were also sterile because they used the rescue splice site. Attempts to rescue males with alternate expression arrangements were not successful, though some male homozygotes had less severe intersex phenotypes. Though this negatively impacted the drive, models showed that it still had significantly improved suppressive power. Therefore, this design may have wide applicability to dsx-based suppression gene drives in a variety of organisms with intermediate homing drive performance.