Quantifying the Structural and Energetic Consequences of EXOSC3 S1 Domain Variants from a Comparative Assessment of {lambda}-Dynamics with Two Charge-Changing Perturbation Strategies
Quantifying the Structural and Energetic Consequences of EXOSC3 S1 Domain Variants from a Comparative Assessment of {lambda}-Dynamics with Two Charge-Changing Perturbation Strategies
Barron, M. P.; Wijeratne, H. R. S.; Runnebohm, A. M.; Caric, K. M.; Mosley, A. L.; Vilseck, J. Z.
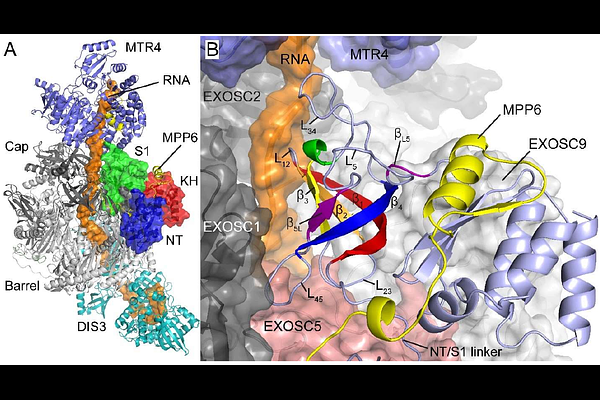
AbstractCharge-changing perturbations are notoriously difficult to investigate with alchemical free energy calculations. The routine use of periodic boundary conditions and electrostatic approximations, such as particle-mesh Ewald (PME), may produce finite-size effect errors that become non-negligible as a perturbation changes a simulation cell\'s net charge away from zero. Two prevalent strategies exist to correct for these errors: the analytic correction (AC) and co-alchemical ion (CI) methods. Both correction schemes have been found to produce comparable relative free energy results for small molecule perturbations, but these methods have not been compared using {lambda}-dynamics ({lambda}D) free energy calculations or for protein side chain mutations. Recently, we investigated relative folding and binding free energies ({Delta}{Delta}Gs) of a series of EXOSC3 variants involved in a rare neurodegenerative disorder, including D132A, G135R, and G191D charge-change perturbations, with a simplified AC scheme in {lambda}D. In this study, these perturbations are reevaluated with the CI scheme for comparison with AC to identify the best correction strategy for {lambda}D. The collected AC- and CI-corrected {Delta}{Delta}Gs show excellent agreement with a mean unsigned error of 0.4 kcal/mol. However, reduced sampling proficiency and increased difficulties of evaluating multisite perturbations with the CI method suggest that a simplified AC approach may be more generalizable for future {lambda}D calculations. Previously, the use of the CI approach with {lambda}D has been limited due to a lack of infrastructure available to users to simplify its more involved setup procedure. This study introduces an automated workflow for implementation of the CI approach with {lambda}D, laying the foundation for future comparisons between charge-change correction schemes. These studies facilitated analysis of the {lambda}D trajectories to identify structural changes within EXOSC3 and the RNA exosome complex that clearly rationalize the calculated {Delta}{Delta}Gs for the D132A, G135R, G191C, and G191D EXOSC3 variants, providing insight into potential disease-causing mechanisms of EXOSC3 modifications.