Holotype genome of the lesula provides insights into demography and evolution of a threatened primate lineage
Holotype genome of the lesula provides insights into demography and evolution of a threatened primate lineage
Jensen, A.; Horton, E. R.; Bisimwa, K.; Detwiler, K. M.; Guschanski, K.
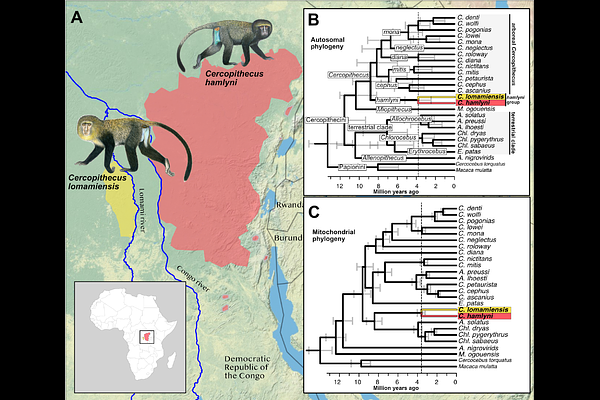
AbstractThe rapid development of genome sequencing techniques in recent decades has revolutionized the field of evolutionary biology, facilitating the study of adaptation and speciation at the genome level. Genomic data has also become a cornerstone in conservation management, allowing inferences of population demography and genetic diversity. Here, we sequenced the genome of the holotype specimen of the elusive lesula (Cercopithecus lomamiensis), a recently described member of the guenons (tribe Cercopithecini) endemic to the Democratic Republic of the Congo. Using published and novel genomic data, we explore the evolutionary and demographic history of C. lomamiensis and its sister species C. hamlyni, the only two members of the hamlyni species group. We estimate that the two species split ca. 3-4 million years ago, and find that they both show high genetic diversity despite being listed as vulnerable on the IUCN Red List. We identify signatures of positive selection in the hamlyni group in genes involved in, e.g., pelage coloration and immune functions, as well as skeletal morphology and locomotor behavior, potentially related to their terrestrial lifestyle which stands out among the otherwise arboreal Cercopithecus genus. We specifically explored whether introgression from more distantly related terrestrial guenons was involved in the evolution of terrestriality in the hamlyni group. However, molecular convergence with other terrestrial species was low, suggesting that putative terrestrial adaptations occurred largely independently. By sequencing the C. lomamiensis genome, this study provides insights into the demography and evolutionary history in a threatened primate lineage, with relevance to conservation management.