Analysis of protein aging reveals rates of subcellular organelle renewal and selective post-translational modification in Arabidopsis
Analysis of protein aging reveals rates of subcellular organelle renewal and selective post-translational modification in Arabidopsis
Tivendale, N.; Liu, X.; Millar, A. H.
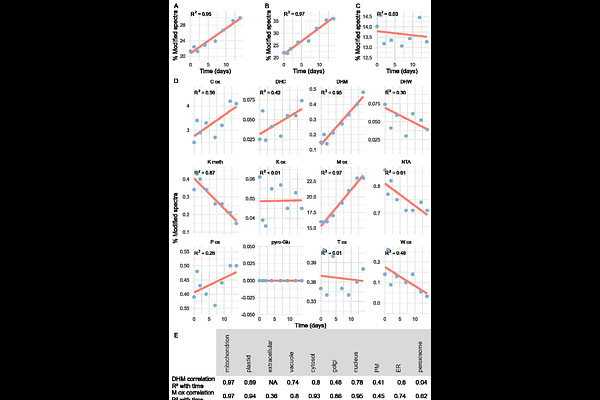
AbstractThe cellular proteome represents a mixture of older and newer copies of each protein type and turnover of this mixture occurs by cycles of protein synthesis and degradation. There is considerable research on new protein synthesis, the nature of nascent proteins and cellular machinery of protein degradation. However, we have limited insights into older proteins or the protein aging process in plants at scale. In this study we use pulse chase biorthogonal non-canonical amino acid tagging (BONCAT) in Arabidopsis cells coupled to affinity purification to capture and analyse snapshots of the cellular proteome as it ages over a two-week period. Each snapshot was subjected to peptide mass spectrometry-based identification, quantitation and characterisation. We show that there are a broad range of lifespans among the 1688 proteins studied and that their subcellular location correlates strongly with protein longevity. Mitochondria, plastids and the extracellular environment contained the longest lived sub-proteomes while the vesicular pathway to ER, PM and peroxisomes contained the shortest-lived protein sets. Abundant primary metabolic enzymes have considerable longevity, while kinases and ubiquitination machinery do not. Through analysis of the aging profiles, we demonstrate that many proteins selectively accumulate posttranslational modifications (PTMs) as they age and that these are mostly oxidative in nature. We show by analysis of exemplar proteins that distinct PTM profiles and proportional changes with age exist between proteins, likely dictated by differences in subcellular environment and protein function. Implications of these insights for understanding cellular function and for biotechnological modification of the plant proteome are discussed.