Conformational Landscaping and Dynamic Mutational Profiling of Binding Interactions and Immune Escape for Broadly Neutralizing Class I Antibodies with SARS-CoV-2 Spike Protein: Distributed Binding Hotspot Networks Underlie Mechanism of Viral Resistance Against Existing Variants
Conformational Landscaping and Dynamic Mutational Profiling of Binding Interactions and Immune Escape for Broadly Neutralizing Class I Antibodies with SARS-CoV-2 Spike Protein: Distributed Binding Hotspot Networks Underlie Mechanism of Viral Resistance Against Existing Variants
Alshahrani, M.; Parikh, V.; Foley, B.; Verkhivker, G.
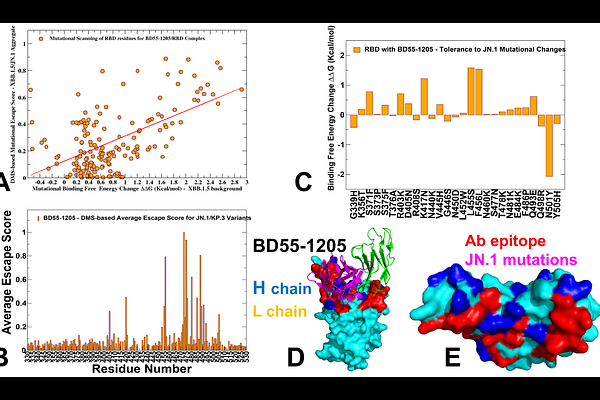
AbstractThe rapid evolution of SARS-CoV-2 has underscored the need for a detailed understanding of antibody binding mechanisms to combat immune evasion by emerging variants. In this study, we investigated the interactions between Class I neutralizing antibodies BD55-1205, BD-604, OMI-42, P5S-1H1, and P5S-2B10 and the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein using multiscale modeling which combined coarse-grained simulations and atomistic reconstruction of conformational landscapes together with mutational scanning of the binding interfaces, dynamic profiling of binding and immune escape using molecular mechanics generalized Born surface area (MM-GBSA) analysis. A central theme emerging from this work is the critical role of epitope breadth and interaction diversity in determining an antibody resilience to mutations. BD55-1205 antibody exemplifies the advantages of broad epitope coverage and distributed hotspot mechanisms. By engaging an extensive network of residues across the RBD, BD55-1205 minimizes its dependence on individual side-chain conformations, allowing it to maintain robust binding even when key residues are mutated. This adaptability is particularly evident in its tolerance to mutations at positions such as L455 and F456 , which severely compromise other antibodies. The ability of BD55-1205 to sustain cumulative interactions underscores the importance of targeting diverse epitopes through multiple interaction mechanisms, a strategy that enhances resistance to immune evasion while maintaining functional integrity. In contrast, BD-604 and OMI-42, with localized binding mechanisms, are more vulnerable to escape mutations at critical positions such as L455, F456, and A475. P5S-1H1 and P5S-2B10 exhibit intermediate behavior, balancing specificity and adaptability but lacking the robustness of BD55-1205. Mutational scanning identified key residues Y421, Y489, and F456 as critical hotspots for RBD stability and antibody binding, highlighting their dual role in viral fitness and immune evasion. The computational predictions generated through mutational scanning and MM-GBSA analysis demonstrate excellent agreement with experimental data on average antibody escape scores. This study underscores the diversity of binding mechanisms employed by different antibodies and molecular basis for high affinity and excellent neutralization activity of the latest generation of antibodies.