Structural basis of RNA-guided transcription by a dCas12f-σE-RNAP complex
Structural basis of RNA-guided transcription by a dCas12f-σE-RNAP complex
Xiao, R.; Hoffmann, F. T.; Xie, D.; Wiegand, T.; Palmieri, A. I.; Sternberg, S. H.; Chang, L.
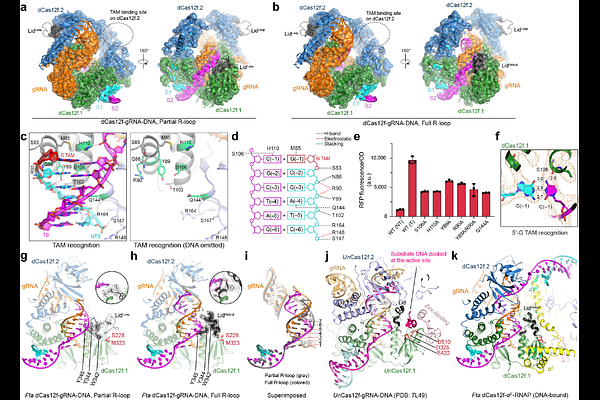
AbstractRNA-guided proteins have emerged as critical transcriptional regulators in both natural and engineered biological systems by modulating RNA polymerase (RNAP) and its associated factors(1-5). In bacteria, diverse clades of repurposed TnpB and CRISPR-associated proteins repress gene expression by blocking transcription initiation or elongation, enabling non-canonical modes of regulatory control and adaptive immunity(1,6,7). Intriguingly, a distinct class of nuclease-dead Cas12f homologs (dCas12f) instead activates gene expression through its association with unique extracytoplasmic function sigma factors ({sigma}E)(8), though the molecular basis has remained elusive. Here we reveal a novel mode of RNA-guided transcription initiation by determining cryo-electron microscopy structures of the dCas12f-{sigma}E system from Flagellimonas taeanensis. We captured multiple conformational and compositional states, including the DNA-bound dCas12f-{sigma}E-RNAP holoenzyme complex, revealing how RNA-guided DNA binding leads to {sigma}E-RNAP recruitment and nascent mRNA synthesis at a precisely defined distance downstream of the R-loop. Rather than following the classical paradigm of {sigma}E-dependent promoter recognition, these studies show that recognition of the -35 element is largely supplanted by CRISPR-Cas targeting, while the melted -10 element is stabilized through unusual stacking interactions rather than insertion into the typical recognition pocket. Collectively, this work provides high-resolution insights into an unexpected mechanism of RNA-guided transcription, expanding our understanding of bacterial gene regulation and opening new avenues for programmable transcriptional control.