Nucleation Landscape of Biomolecular Condensates in the Grand Canonical Ensemble via Monte Carlo Simulations
Nucleation Landscape of Biomolecular Condensates in the Grand Canonical Ensemble via Monte Carlo Simulations
Sepehri, A.; Zerze, G. H.
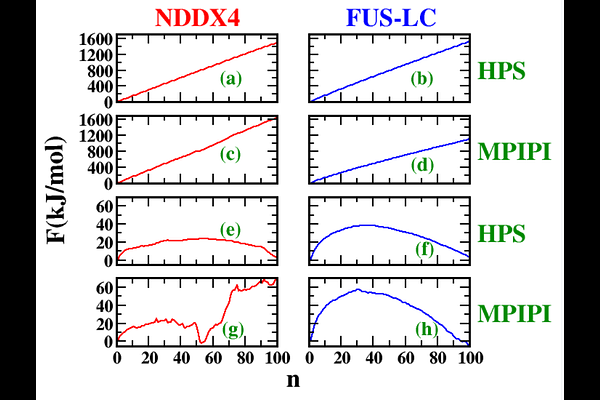
AbstractWe present a Monte Carlo framework for simulating the nucleation of biomolecular condensates in the grand canonical ensemble, which overcomes the limitations of fixed particle number and allows direct control of the dilute-phase concentration. Our approach combines conformation sampling with bias-enhanced cluster size sampling, enabling accurate sampling of individual cluster sizes in various conformations. Our method resolves nucleation free energy surfaces and is capable of capturing both classical and nonclassical nucleation mechanisms. We validated our method by reproducing structural properties of disordered proteins across a diverse benchmark set and by applying it to study nucleation in two phase-separating proteins, FUS-LC and NDDX4, using both HPS and MPIPI coarse-grained force fields. While both proteins exhibit classical nucleation behavior under the HPS model, only FUS-LC remains classical with MPIPI. In contrast, NDDX4 shows a distinctly nonclassical nucleation pathway under MPIPI, characterized by a metastable intermediate state near the cluster size of 53 protein chains. Morphological analysis reveals that clusters up to this point are compact and spherical, whereas larger clusters adopt a nonspherical, two-lobed geometry indicative of frustrated coalescence. These findings underscore the critical role of sequence composition and force field parameterization in shaping nucleation pathways and demonstrate the utility of our framework for uncovering complex, mechanism-rich free energy landscapes in biomolecular condensation.