Enhancement of hidden Markov model analyses for improved inference of archaic introgression in modern humans
Enhancement of hidden Markov model analyses for improved inference of archaic introgression in modern humans
Coll Macia, M.; Skov, L.; Baek, Z. E. D.; Hobolth, A.
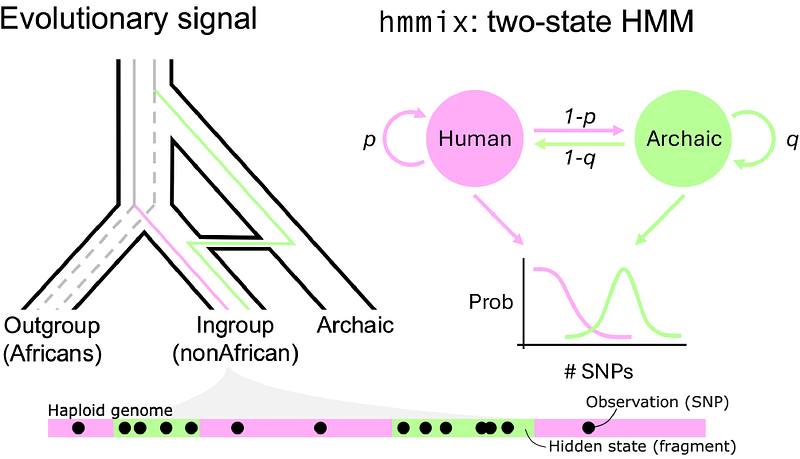
AbstractInsights into the admixture history between modern and archaic humans require accurately inferred introgressed fragments within modern genomes. Here, we introduce two enhancements to hidden Markov models (HMMs) implemented in hmmix. First, we develop a method for sampling hidden state sequences conditional on observed genomic data, enabling robust estimation of admixture summary statistics - such as admixture proportion and fragment length distributions. This represents an improvement compared to relying solely on point estimates as provided by classical decoding methods. Additionally, we integrate the Finite Markov Chain Imbedding (FMCI) framework, allowing exact analytical calculation of these admixture statistics, tailored to large scale human genomes. Second, we implement a novel hybrid decoding method which combines the strengths of Viterbi and Posterior decoding methods, substantially improving the reliability of archaic fragments identified. We validate these improvements on data from the 1000 Genomes Project and demonstrate that our sampling method yield more accurate admixture estimates from single individuals compared to existing approaches requiring extensive population-level datasets. Moreover we show how hybrid decoding can be instrumental in resolving the inference of local archaic haplotype structure in modern human genomes. These methodological advancements broadly enhance HMM-based analyses and will provide deeper insight into the complex history of genetic interactions between archaic and modern human populations.