Asymmetrical recognition and processing of double-strand breaks formed during DNA replication
Asymmetrical recognition and processing of double-strand breaks formed during DNA replication
Johnson, M. J.; Kimble, M. T.; Jeong, S.; Sane, A.; Symington, L. S.
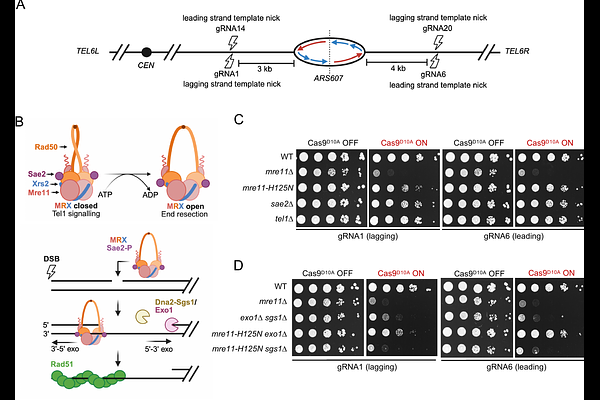
AbstractDNA end resection to generate 3\' ssDNA overhangs is the first step in homology-directed mechanisms of double-strand break (DSB) repair. While end resection has been extensively studied in the repair of endonuclease-induced DSBs, little is known about how resection proceeds at DSBs generated during DNA replication. We previously established a system to generate replication-dependent double-ended DSBs at the sites of nicks induced by the Cas9D10A nickase in the budding yeast genome. Here, we suggest that these DSBs form in an asymmetric manner, with one break end being blunt or near blunt, and the other bearing a 3\' ssDNA overhang of up the size of an Okazaki fragment. We find that Mre11 preferentially binds blunt ends and is required for the removal Ku from these DSB ends. In contrast, the ends predicted to have 3\' overhangs have minimal Ku binding, and end resection at these break ends can proceed in a mostly Mre11-independent manner through either the Exo1 or Dna2-Sgs1 long-range resection pathways. These findings indicate that resection proceeds differently at replication-dependent DSBs than at canonical DSBs, and reveals that Ku selectively binds nearly blunt ends, potentially explaining why replication-dependent DSBs are poorly repaired by non-homologous end joining.