CGBack: Diffusion Model for Backmapping Large-Scale and Complex Coarse-Grained Molecular Systems
CGBack: Diffusion Model for Backmapping Large-Scale and Complex Coarse-Grained Molecular Systems
Ugarte La Torre, D.; Sugita, Y.
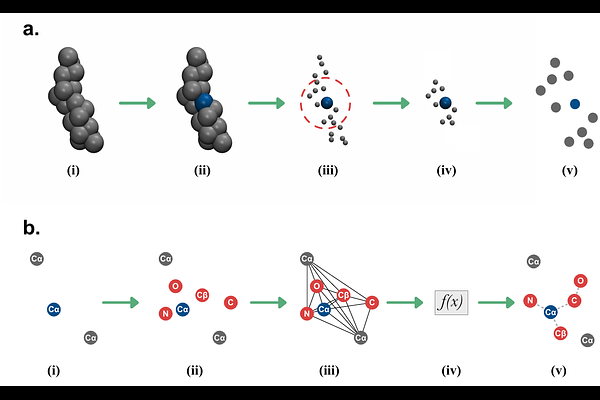
AbstractMolecular dynamics simulations based on coarse-grained (CG) models are used to accelerate conformational dynamics of biomolecules and other chemical systems with reduced computational costs. CG models achieve it by discarding atomic information necessary for downstream structural analysis. Recovering the atomic detail from CG structures, i.e. backmapping, remains a fundamental challenge in multiscale modeling, especially for proteins and complex biomolecular assemblies. Recent machine learning methods have shown promise in reconstructing atomistic details, but most approaches only target simple systems at a small scale. In particular, conventional backmapping pipelines often fail to preserve stereochemistry, induce high-energy configurations, or require extensive minimization. Here we present CGBack, a backmapping framework that employs a denoising diffusion probabilistic model to reconstruct all-atom molecular structures from CG representations. CGBack consists of backmapping and refinement procedures and accurately recovers atomic coordinates across diverse protein systems from small to large scales. We show that CGBack is capable of accurately backmapping both single-chain and multi-chain molecular systems, including densely packed intrinsically disordered proteins in condensates. These results suggest that CGBack can be a powerful tool for multiscale molecular simulation pipelines. We anticipate that CGBack will enable more efficient workflows for protein modelling, as well as for other biomolecules across different CG models.