Evolution and comparative genomics of tick-associated <Francisella> endosymbionts: insights into metabolic pathways and historical biogeographic patterns
Evolution and comparative genomics of tick-associated <Francisella> endosymbionts: insights into metabolic pathways and historical biogeographic patterns
Echeverry-Perez, J. S.; Castelli, M.; Munoz-Leal, S.; Nava, S.; Sassera, D.; Sanchez-Vialas, A.; Olmeda, A. S.; Valcarcel, F.; Uribe, J. E.
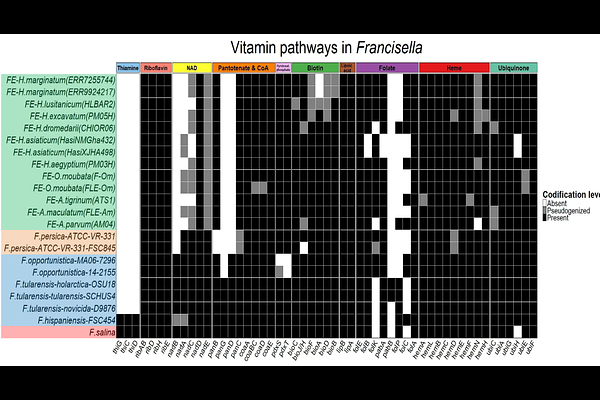
AbstractTicks (Ixodida) are the second most important vectors of infectious diseases in vertebrates, after mosquitoes. They also maintain mutualistic relationships with bacteria, such as endosymbionts that provide essential B vitamins absent in their blood diet. The most studied endosymbionts belong to the genera <Coxiella>, <Midichloria>, and <Francisella>. <Francisella> includes endosymbionts (FE), pathogens (FP), putative intermediates (FI), and free-living (FL) strains, making them valuable for evolutionary and comparative genomics. In this study, total DNA from six adult female ticks of the genera <Hyalomma> and <Amblyomma> was sequenced to obtain new FE genomes. Additionally, two deep metagenomes from public data were assembled, resulting in a dataset of 22 <Francisella> strains. This dataset was used to reconstruct a phylogenomic framework and compare vitamin biosynthesis and virulence pathways. An MLST-based dense phylogeny was also built to explore biogeographic patterns. The resulting phylogenomic tree shows FE form a monophyletic group derived from FP, possibly due to historical biogeography or recent horizontal transfers. Comparative analyses reveal that FE retain key metabolic pathways while losing nonessential ones, reflecting a selective genome reduction. These results advance our understanding of symbiont evolution in a changing world, revealing molecular adaptations that underpin tick-symbiont relationships and offering genomic insights with potential applications for disease control.