Cell cycle-coupled transcriptional network orchestrates human B cell fate bifurcation
Cell cycle-coupled transcriptional network orchestrates human B cell fate bifurcation
Pease, N. A.; Fan, J.; Keshari, S.; Stratton, J.; Gerges, P.; Ann Varghese, B.; Nampoothiri VP, N.; McGinnnis, C. S.; Zhang, W.; Geirlack, S. B.; Swaminathan, T.; Sachan, A.; Manakkat Vijay, G.; Mena Hernandez, L.; Heidari Rarani, Z.; Macedo, C.; Metes, D.; Satpathy, A. T.; Jain, A. K.; Sahni, N.; Stallaert, W.; Das, J.; Singh, H.
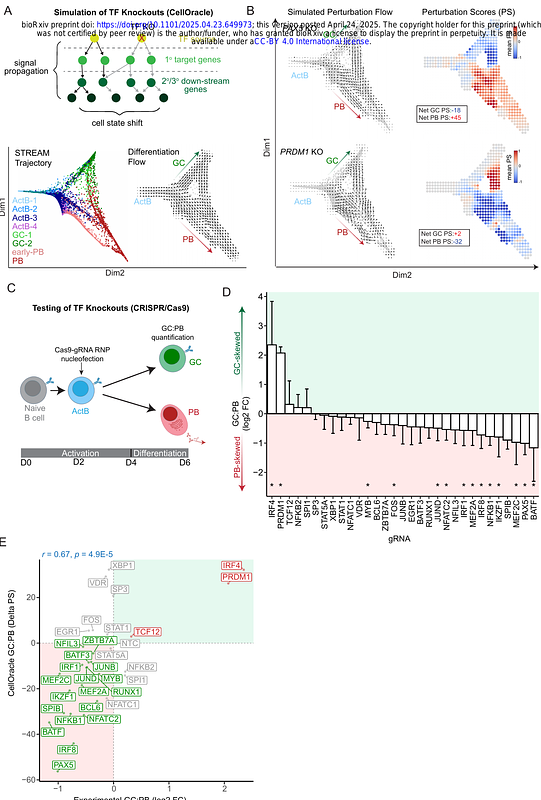
AbstractAntibody responses are determined by activated B cells bifurcating into plasmablasts (PBs) and germinal center B cells (GCBCs). Gene regulatory networks (GRNs) underlying human B cell fate choice remain uncharted. Temporally resolved single-cell multi-omics, computational modeling and CRISPR-based perturbations were used to assemble, simulate and test high-resolution GRNs underlying PB and GC fates. The results converged with orthogonal predictions of transcription factor (TF) action at single-nucleotide resolution, revealing dominant and reciprocal actions of IRF4 and its binding partners at simple and composite IRF motifs. Single-cell perturbation analysis of these TFs demonstrated multiple reciprocal negative feedback loops controlling the bifurcation. Additionally, IRF4 and BLIMP1, co-repressed the cell cycle regulators MYC and CCND2. G0/G1 lengthening accelerated the switching of cells to an IRF4hiBLIMP1hi regulatory state and enhanced the probability of PB specification, thereby uncovering a self-reinforcing regulatory module that couples cell cycle dynamics to B cell fate choice.