Validation of Duchenne muscular dystrophy candidate modifiers using a CRISPR-Cas9-based approach in zebrafish
Validation of Duchenne muscular dystrophy candidate modifiers using a CRISPR-Cas9-based approach in zebrafish
Lerma, G.; Ryhlick, K. R.; Carraher, O. M.; Beljan, J. C.; Amacher, S. L.
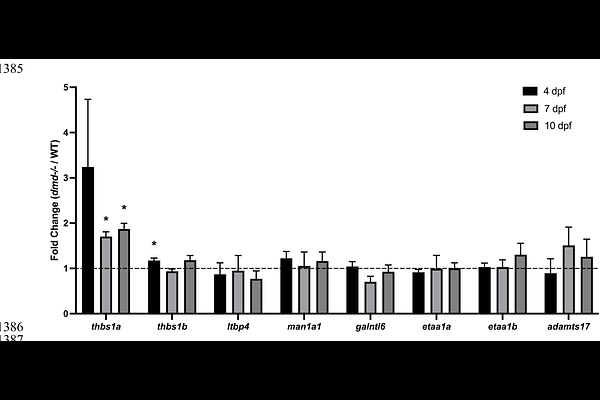
AbstractDuchenne muscular dystrophy (DMD) is a progressive muscle wasting disease for which there is no cure. There is a critical need for additional therapeutics. Human genome-wide association studies (GWAS) have identified candidate DMD genetic modifiers that could serve as therapeutic targets. Because many GWAS-identified single nucleotide polymorphisms (SNPs) lie in noncoding, putative regulatory regions, it can be challenging to identify which gene(s) are regulated by these SNPs and how gene expression is altered to modify disease severity even with extensive in silico modeling. We analyzed expression of zebrafish orthologs of putative DMD modifiers and showed almost all are comparably expressed in wild-type and dmd mutant zebrafish at three different stages of disease. To model decreased expression of candidate modifiers, we pursued a zebrafish CRISPR-based screening approach, which we validated by testing zebrafish orthologs of two extensively studied DMD modifiers, LTBP4 and THBS1. We then tested candidates from the most recent GWAS and demonstrate that galntl6, man1a1, etaa1a;etaa1b, and adamts17 are bona fide DMD modifiers. Our findings demonstrate the utility of zebrafish for DMD genetic modifier screening and characterizing modifier function.