DeepCryoRNA: deep learning-based RNA structure reconstruction from cryo-EM maps
DeepCryoRNA: deep learning-based RNA structure reconstruction from cryo-EM maps
Li, J.; Chen, S.-J.
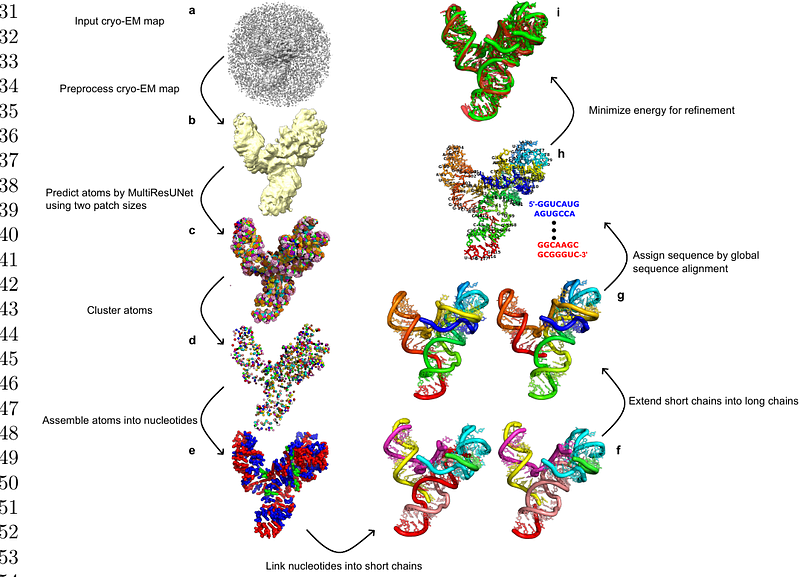
AbstractRNA molecules play a crucial role in various cellular functions, making understanding their three-dimensional (3D) structures vitally important. Cryogenic electron microscopy (cryo-EM) has significantly advanced the study of RNA structures. However, most existing modeling tools are primarily developed for proteins, making them less effective for RNAs due to the unique structural variability and flexibility of an RNA. Addressing the limitations of current cryo-EM based RNA structure determination, we develop DeepCryoRNA, a novel deep learning approach tailored for automated reconstruction of RNA 3D structures from cryo-EM maps at high to medium resolutions. The approach was rigorously evaluated using a benchmark set of 51 RNA structures, resolved by cryo-EM at resolutions finer than 6.0 {\\AA}. The results demonstrate DeepCryoRNA\'s marked accuracy over existing methods. The methodology of DeepCryoRNA stands out for its accurate atomistic predictions and advanced global sequence alignment capabilities, enabling more accurate sequence assignments. These features collectively establish DeepCryoRNA as a highly efficient and effective tool for RNA 3D structure reconstruction from cryo-EM maps, advancing the field of RNA structural biology.