Systematic non-natural base-pairs for strand displacement circuits in complex environments
Systematic non-natural base-pairs for strand displacement circuits in complex environments
Kennedy, T. J.; Mayer, T.; Simmel, F.; Thachuk, C.
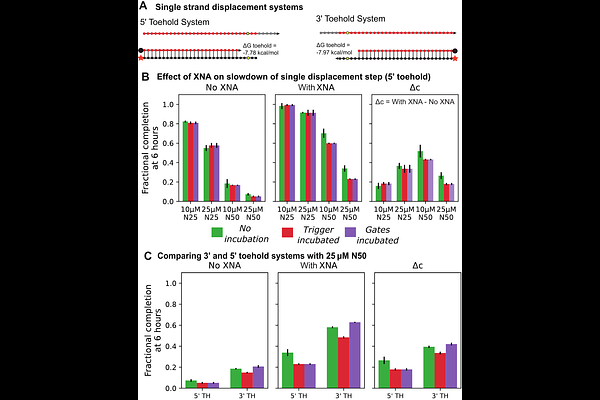
AbstractNucleic acid strand displacement (SD) circuits have enabled impressive realizations of molecular computing in vitro, but operating them in contention with competing reaction pathways can cause signal leak and circuit failure. Further, operating SD circuits in complex environments, ranging from pools of nucleic acids to living organisms, can exacerbate these challenges. To address these issues, we explore the incorporation of isoC and isoG non-natural bases in single- and multi-step SD cascades and started by studying their promiscuity. We combined the non-natural bases with a mismatch strategy, showing them to be fully compatible. The synthesis we show between strategic placement of non-natural bases and systematic mismatch optimization of SD circuits offers a design space for future circuits that are fast, robust, and biologically compatible. Comparing circuit slowdowns by random pool backgrounds in the presence and absence of non-natural base pairs reveals a significant difference. Using isoC:isoG base pairs in the toehold region shows the greatest effect, with some further positive effects caused by additional non-natural bases in the branch migration domain. Such insights will pave the way for developing SD circuits that operate in complex environments, expanding the applications of molecular computing in a wider range of scenarios.