Assembly of Macromolecular Complexes in the Whole-Cell Model of a Minimal Cell
Assembly of Macromolecular Complexes in the Whole-Cell Model of a Minimal Cell
Fu, E.; Thornburg, Z. R.; Brier, T. A.; Wei, R.; Bo, Y.; Gilbert, B. R.; Wang, S.; Luthey-Schulten, Z.
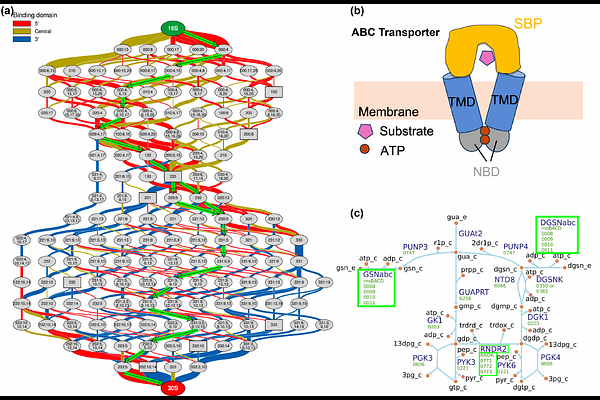
AbstractMacromolecular complexes in the genetically minimized bacterium, JCVI-syn3A, support gene expression (RNA polymerase, ribosome, degradosome), metabolism (ABC transporters, ATP synthase) and chromosome dynamics. In this work, we further incorporate the assembly of 21 unique macromolecular complexes into the existing whole-cell kinetic model of Syn3A. The synthesis and translocation of protein subunits in membrane complexes occur through distinct pathways. A range of assembly rates were considered to guarantee a high yield of assembly given the existing time scales of the gene expression with derived formula to rationalize the choice. By alleviating the undesired dead-end intermediates in ATP synthase assembly, the efficiency was improved, while the heterogeneity of subunit synthesis at the single-cell level greatly undermine it. The assembly of RNA polymerase, ribosome, and degradosome influence the speed and efficiency of protein synthesis. Collectively, this model predicted time-dependent cellular behaviors consistent with experiments. A deep learning analysis of the simulated time-dependent concentrations of 148 intracellular metabolites allowed the construction of a trajectory tree from which three distinct metabolic phenotypes were identified in the population of ~ 100 cells considered.