Prediction of physical characteristics of disordered proteins using molecular simulation and physics-informed multiple machine learning strategies
Prediction of physical characteristics of disordered proteins using molecular simulation and physics-informed multiple machine learning strategies
Gonzalez, D. L.; Ibrahim, S.; Atia, G.; Seth, S.; Bhattacharya, A.
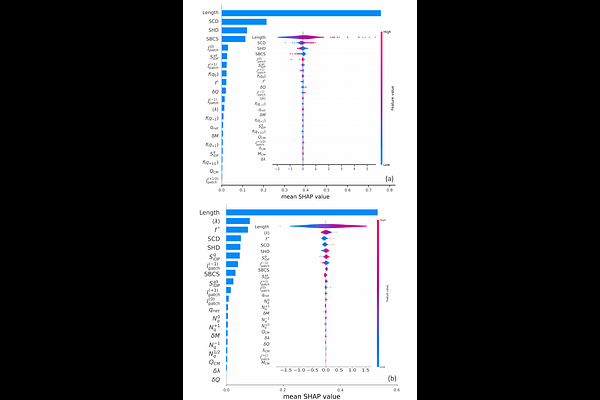
AbstractWe report novel Machine Learning (ML) strategies to predict gyration radii and other physical characteristics of intrinsically disordered proteins (IDPs) by introducing a novel hybrid architecture. Our architecture combines a sequence-based deep neural network (DNN) that processes IDP sequence information with a fully connected neural network (FCNN) using a set of physical features drawn from a coarse-grained (CG) model of IDPs validated by experimental data. Brownian dynamics (BD) simulation results for the gyration radii and other physical characteristics for approximately 7000 IDPs drawn from MobiDB database with > 99% disorder score and the characteristics 23 physical features of the CG model were used to train several explainable Machine Learning (ML) models (FCNN, RNN, GRU, biGRU, LSTM, and BiLSTM) to predict conformational properties of IDPs. We conclude that the sequence-based models outperformed feature-only models on the test data, with the GRU model achieving the best performance among sequence-only approaches. In general we find that combining sequence-based and feature-based information provides better accuracy for all architectures with Hybrid biGRU model achieving the best predictive performance among all tested architectures. The SHAP analysis for all the architectures reveal relative importance of different molecular features and provide routes to narrow down the MSE. Furthermore an integrated analysis reveals that apart from the length of the IDPs, the three parameters (SCD, SHD, and f* play key role in ML predictions. Our method provides an excellent platform for fast and accurate prediction of conformation properties for the new IDPs for rapid initial screening and exploration of novel properties which further then could be studied using more expensive molecular simulation.