Planetary-scale metagenomic search reveals new patterns of CRISPR targeting
Planetary-scale metagenomic search reveals new patterns of CRISPR targeting
Roux, S.; Neri, U.; Bushnell, B.; Fremin, B.; George, N.; Gophna, U.; Hug, L. A.; Camargo, A. P.; Wu, D.; Ivanova, N.; Kyrpides, N. C.; Eloe-Fadrosh, E. A.
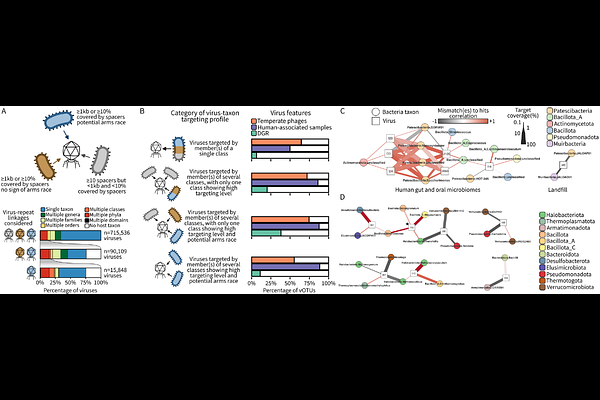
AbstractInteractions between microbes and their mobile genetic elements (MGEs), including viruses and plasmids, are critical drivers of microbiome structures and processes. CRISPR-Cas systems are known to be important regulators of these host-MGE interactions, but a global understanding of CRISPR-Cas diversity, activity, and roles across Earth\'s biomes is still lacking. Here, we use an optimized computational approach to search short-read data and collect ~800 million CRISPR spacers across ~450,000 public metagenomes. Comparing spacers across samples and taxa revealed a high population diversity for CRISPR loci overall, with typically only a small subset of spacers detected as prevalent and conserved within a microbial population. From this extensive CRISPR spacer dataset, we identified 1.18 billion hits between 41 million spacers and 2.5 million viruses and plasmids. Prevalent and conserved spacers were over-represented in these MGE-matching spacers, and CRISPR spacers frequently matched multiple MGEs, consistent with a positive selection pressure associated with MGE targeting. Focusing on the role of CRISPR as anti-phage defense, we observed surprising cases of viruses targeted by microbes not expected to be viable hosts. These were more frequent for viruses encoding a diversification mechanism (DGR) for their host attachment proteins, and associated with a reduced rate of escape mutations. This suggests that broad spacer targeting may derive from the recurring entry of a virus genome into a non-host microbial cell, leading to some viruses being targeted by taxonomically diverse microbes well outside of their actual host range. Taken together, this petabase-scale exploration of CRISPR arrays in nature outlines the extensive diversity of CRISPR array loci across microbiomes. It highlights several key genomic and ecological parameters driving the activity of CRISPR arrays that are likely influencing strain-level diversification and selection processes within microbial populations.