Direct observation of Notch signalling induced transcription hubs mediating gene-expression responses
Direct observation of Notch signalling induced transcription hubs mediating gene-expression responses
Santa-Cruz Mateos, C.; Roussos, C.; de Haro Arbona, F. J.; Falo Sanjuan, J.; Bray, S.
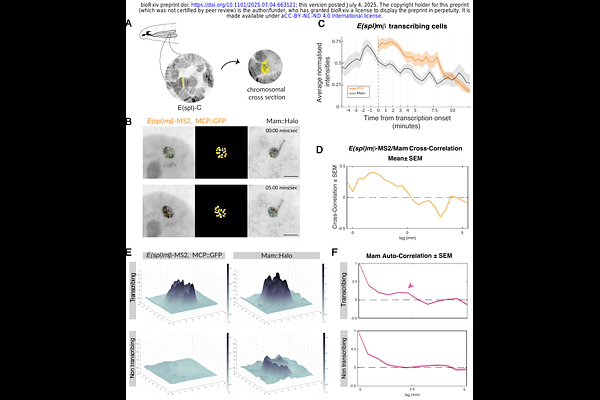
AbstractDevelopmental decisions rely on cells making accurate transcriptional responses to signals they receive, as with Notch pathway activity. Local condensates or transcription factor hubs are a proposed mechanism for facilitating gene activation by nuclear complexes. To investigate their importance in endogenous Notch signalling, we deployed multi-colour live-imaging to measure Notch transcription-complex enrichment at a target gene locus in combination with the transcription dynamics. The co-activator Mastermind (Mam) was present in signalling-dependent nuclear foci, during Notch active developmental stages. Tracking these highly-dynamic Mam hubs together with transcription in the same nucleus, revealed that their condensation precedes and correlates with the profile of transcription and becomes stabilized if transcription is inhibited. Manipulations to signalling levels had concordant effects on hub intensities and transcription profiles, altering their probability and amplitude. Together the results argue that signalling induces the formation of transcription hubs whose properties are instrumental in the quantitative gene expression response to Notch activation.