Efficient Estimation of Nucleotide Diversity and Divergence using Depth Information
Efficient Estimation of Nucleotide Diversity and Divergence using Depth Information
Mirchandani, C. D.; Enbody, E.; Sackton, T. B.; Corbett-Detig, R.
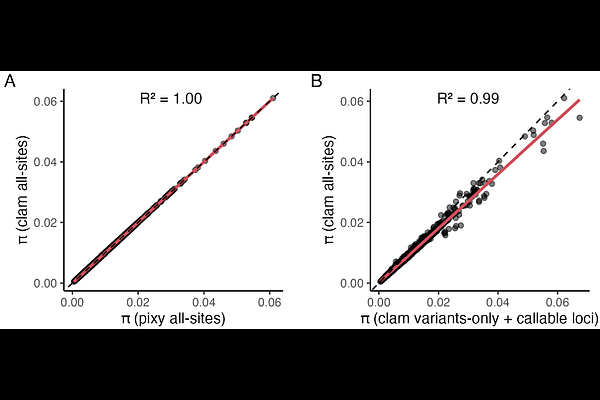
AbstractThe increasing scale of population genomic datasets presents computational challenges in estimating summary statistics such as nucleotide diversity ({pi}) and divergence (dxy). Unbiased estimates of diversity require knowledge of missing data and existing tools require all-sites VCFs. However, generating these files is computationally expensive for large datasets. Here, we introduce Callable Loci And More (clam), a tool that leverages callable loci -- determined from depth information -- to estimate population genetic statistics using a variant-only VCF. This approach offers improvements in storage footprint and computational performance compared to contemporary methods. We benchmark clam using a large muskox dataset and demonstrate that it produces unbiased estimates of {pi} while reducing runtime and storage requirements, compared to an existing approach. clam provides an efficient and scalable alternative for population genomic analyses, facilitating the study of increasingly large and diverse datasets. clam is available as a standalone program and integrated into snpArcher for efficient reproducible population genomic analysis.