Discriminating models of trait evolution
Discriminating models of trait evolution
Lozano, J. R.; DeGiorgio, M.; Assis, R.; Adams, R.
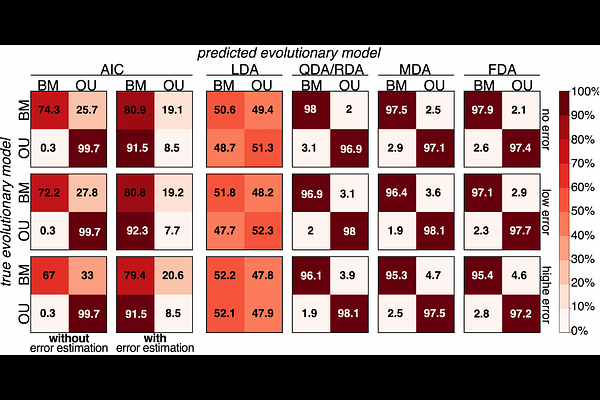
AbstractA central challenge in comparative biology is linking present-day trait variation across species with unobserved evolutionary processes that occurred in the past. In this endeavor, phylogenetic comparative methods are invaluable for fitting, comparing, and selecting evolutionary models of varying complexity and biological meaning. Traditionally, evolutionary studies have relied on conventional statistical approaches to assess model fit and identify the one that best explains variation in a given trait. Here we explore an alternative strategy by applying supervised learning to predict evolutionary models via discriminant analysis. We formally introduce Evolutionary Discriminant Analysis (EvoDA) as an addition to the biologist\'s toolkit, offering a suite of new methods for studying trait evolution. We evaluate the performance of EvoDA alongside conventional model selection through a series of fungal phylogeny case studies, each targeting increasingly challenging analytical tasks. These results showcase the strengths of EvoDA, with substantial improvements over conventional approaches when studying traits subject to measurement error, which likely reflect realistic conditions in empirical datasets. To complement our simulation-based benchmarking, we explore the application of EvoDA for tackling a notoriously difficult task: predicting the mode and tempo of gene expression evolution. This empirical analysis suggests that stabilizing selection acts on a majority of genes, with bursts of expression evolution in a handful of genes related to stress, cellular transportation, and transcription regulation. Collectively, our findings illustrate the promise of EvoDA for predicting trait models across a range of evolutionary and experimental contexts, establishing a new methodological framework for the next era of comparative research.