Personalized whole-brain Ising models with heterogeneous nodes capture differences among brain regions
Personalized whole-brain Ising models with heterogeneous nodes capture differences among brain regions
Craig, A. G.; Chen, S.; Tang, Q.-Y.; Zhou, C.
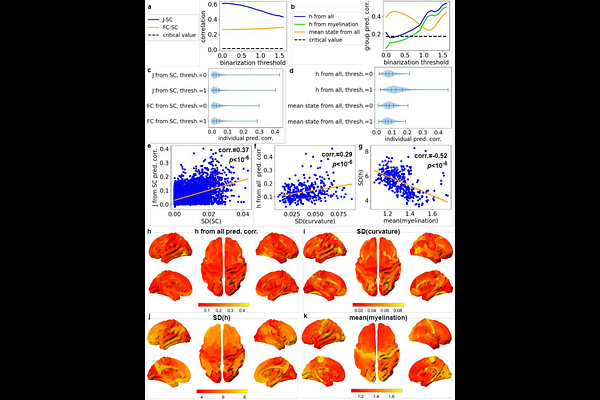
AbstractMultiple lines of research have studied how complex brain dynamics emerge from underlying connectivity by using Ising models as simplified neural mass models. However, limitations on parameter estimation have prevented their use with individual, high-resolution human neuroimaging data. Furthermore, most studies focus only on connectivity, ignoring node heterogeneity, even though real brain regions have different structural and dynamical properties. Here we present an improved approach to fitting Ising models to 360-region functional MRI data: derivation of an initial guess model from group data, optimization of simulation temperature, and two stages of Boltzmann learning, first with group data, then with individual data. We then analyze how data binarization threshold affects goodness-of-fit, the role of the external field in model behavior, and correlations between model parameters and features from structural MRI, including myelination and sulcus depth. The fitted models can reproduce the functional connectivity of the data across a wide range of thresholds, but the role of the external field parameters in model behavior increases with threshold. In parallel, the coupling between nodes correlates with structural connectivity throughout this range, but correlations between the external field and structural features increase with threshold. These results show how our methodology enables personalized, biophysically interpretable modeling of structure-function relationships at the whole-brain level, which can aid understanding of individual differences in brain network organization and dynamics. This approach will help to bridge the gap between connectomics, which emphasizes brain networks, and translational neuroscience, which often focuses on the unique roles of brain regions.