Spatial joint profiling of DNA methylome and transcriptome in mammalian tissues
Spatial joint profiling of DNA methylome and transcriptome in mammalian tissues
Lee, C. N.; Fu, H.; Cardilla, A.; Zhou, W.; Deng, Y.
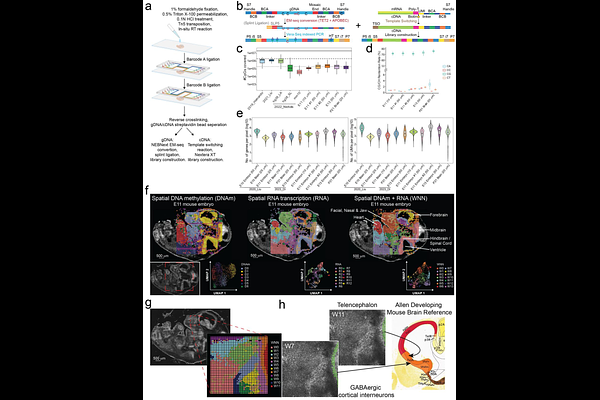
AbstractThe spatial resolution of omics dynamics is fundamental to understanding tissue biology. Spatial profiling of DNA methylation, which is a canonical epigenetic mark extensively implicated in transcriptional regulation, remains an unmet demand. Here, we introduce a method for whole genome spatial co-profiling of DNA methylation and transcriptome of the same tissue section at near single-cell resolution. Applying this technology to mouse embryogenesis and postnatal brain resulted in rich DNA-RNA bimodal tissue maps. These maps revealed the spatial context of known methylation biology and its interplay with gene expression. The two modalities concordance and distinction in spatial patterns highlighted a synergistic molecular definition of cell identity in spatial programming of mammalian development and brain function. By integrating spatial maps of mouse embryos at two different developmental stages, we reconstructed the dynamics of both epigenome and transcriptome underlying mammalian embryogenesis, revealing details in sequence, cell type, and region-specific methylation-mediated transcriptional regulation. This method extends the scope of spatial omics to DNA cytosine methylation for a more comprehensive understanding of tissue biology over development and disease.