Transcription-based identification of uncharacterized genes in the human immune response
Transcription-based identification of uncharacterized genes in the human immune response
Vorsteveld, E. E.; Kersten, S.; Kaffa, C.; Simons, A.; 't Hoen, P. A. C.; Netea, M. G.; Hoischen, A.
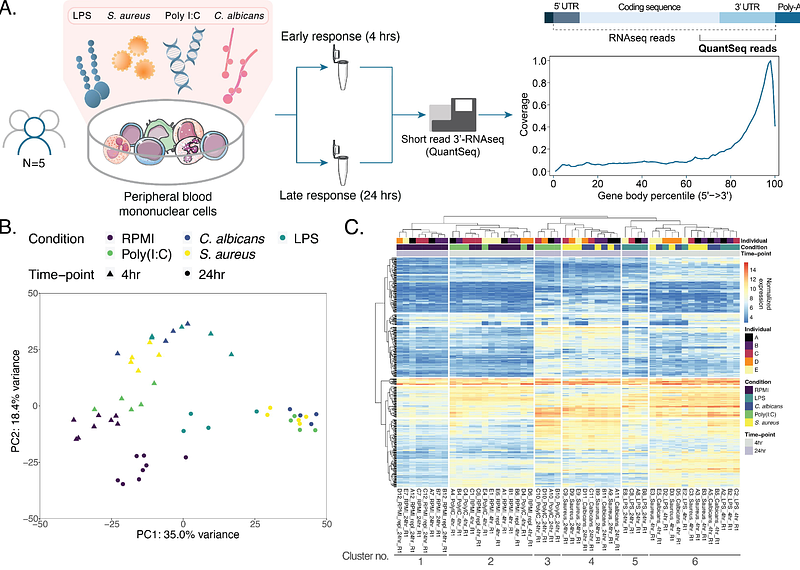
AbstractHost-pathogen interactions are shaped by the nature of the pathogen and by host-related factors. The human host responses can be characterized in microbe-stimulated immune cells using transcriptomics. We set out to characterize gene expression changes as a result of microbial in vitro stimulation in RPMI-cultured human primary immune cells, using four pathogen ligands representing bacteria (LPS and S. aureus), viruses (Poly(I:C)) and fungi (C. albicans) for 4 and 24 hours, resulting in a total of 52 samples for analysis. We analyze the transcriptional changes on gene- and pathway level, highlighting common and distinct effects of pathogen stimulation. We highlight genes without a known function that were differentially expressed as a common effect of pathogen stimulation. We find uncharacterized genes such as KIAA0040 and CYRIA to be co-expressed with genes involved in innate immunity and downstream signaling from the PRRs, respectively. We identify 901 variants in uncharacterized genes in a cohort of patients with rare immune disorders, prioritizing 5 candidate variants in 4 genes that could underly these diseases. Our results therefore indicate the potential role of these genes the human immune response and provides candidate variants with potential relevance in rare immune-mediated diseases.