A pan-generic marker panel for apples to enable genetic research and breeding across Malus species
A pan-generic marker panel for apples to enable genetic research and breeding across Malus species
Sakina, A.; Tegtmeier, R.; Feulner, H.; Hickok, D.; Svara, A.; Cho, P.; Clark, M.; Kilian, A.; Glaubitz, J.; Sun, Q.; Khan, A.
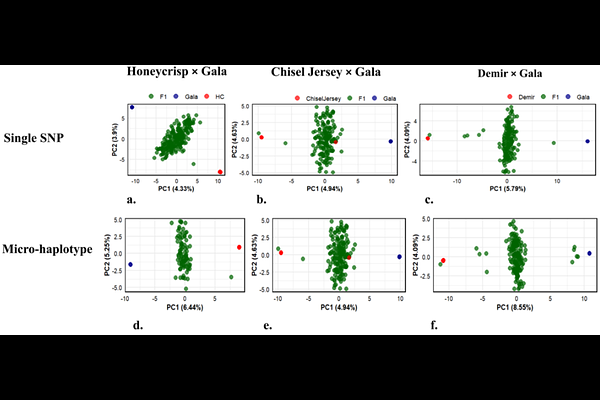
AbstractWild Malus species harbor untapped genetic diversity to advance apple breeding, particularly for disease resistance and stress tolerance. However, existing marker panels, developed mainly using Malus domestica accessions, introduce ascertainment bias and limit detecting rare variants in wild species. We developed and validated a medium-density and cost-effective pan-generic 3K apple DArTag panel optimized to capture genome-wide variation across the Malus genus. The panel was constructed using conserved, syntenic, and collinear genomic blocks identified within the core genome of 13 Malus accessions for cross-species transferability. The panel was validated across three bi-parental mapping populations totaling 593 progeny. Across these populations, 2,461-3,234 SNP markers were polymorphic and 1,482-2,620 were informative. Each population contained over 900 multiallelic micro-haplotype loci, with several hundred loci exhibiting three or four distinct haplotypes. Markers were uniformly distributed across all 17 chromosomes, each containing between 60-230 informative SNPs. The panel was further evaluated on 174 diverse germplasm accessions from 20 Malus species. It exhibited strong cross-species transferability, exceptionally low rates of missing data (<0.5%), and clear genetic differentiation between wild and domesticated accessions. Genome-wide association studies (GWAS) identified a major locus on chromosome 4 significantly linked to fruit length, weight, and width in addition to trait-specific associations on chromosomes 1, 6, 9, and 11. The cost-effectiveness of genotyping per sample (<$15), combined with these results, underscore the panel\'s broad utility for quantitative trait locus (QTL) mapping in bi-parental and diverse populations, marker-assisted selection in the breeding programs, and genetic diversity analysis across the Malus genus.