A rapid and inexpensive universal PCR protocol for DNA (meta)barcoding using a one-tube, 2-step PCR
A rapid and inexpensive universal PCR protocol for DNA (meta)barcoding using a one-tube, 2-step PCR
Collard, O.; Tawfeeq, M.; Ducret, H.; Flot, J.-F.
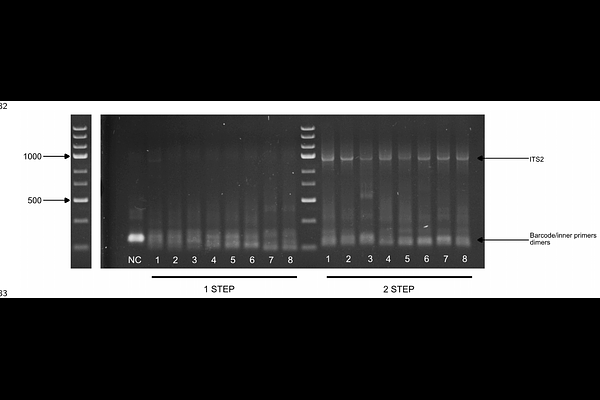
AbstractDNA barcoding has become a widespread technique for species identification through the amplification of standardized gene regions. These include the cytochrome c oxydase subunit I (COI) for metazoans, ITS for fungi and plants or the 16S rRNA for bacteria and archaea. It has been shown to be more accurate than more traditional methods such as morphology, making it a now widespread method for species delimitation. The development of new sequencing methods such as Nanopore sequencing becoming more affordable and accurate has led to a growing use of those methods and a need for fast, standardized and cheap protocol. Although Sanger sequencing has become superseded by next-generation sequencing for most genomic applications, using second or third-generation sequencing for amplicons remains an expensive and time-consuming approach, especially for laboratories that analyse many different markers. Amplicon sequencing typically occurs in the context of either DNA barcoding (sequencing one or a few markers of interest for each specimen in a collection) or DNA metabarcoding (investigating the composition of a community by using universal primers targeting a whole group of organisms). Pooling the resulting PCR products for e.g. Nanopore sequencing requires the use of barcodes that are generally added to amplicons once they are amplified (using either ligation or a second round of amplification), and these supplementary steps are time-consuming, expensive and prone to contamination. To alleviate these issues, we present here a quick, single-tube two-step PCR with four primers: two internal primers with tails, and two external barcode primers that anneal on the tails during the second phase of the PCR. we show an improved 4 primers PCR protocol by using a single tube \"drop in the lid\" method. 8 DNA extract from Asellus. sp were obtained and amplified using both, 2-step PCR and the 1-step PCR. Results shows a clear enhancement of PCR product using the 2-step PCR for both ITS and CO1 marker compare to the 1-Step protocol.