Adding 3Di characters to amino acid datasets can improve resolution, but the effect is weaker in shorter and alpha-helical proteins such as histones
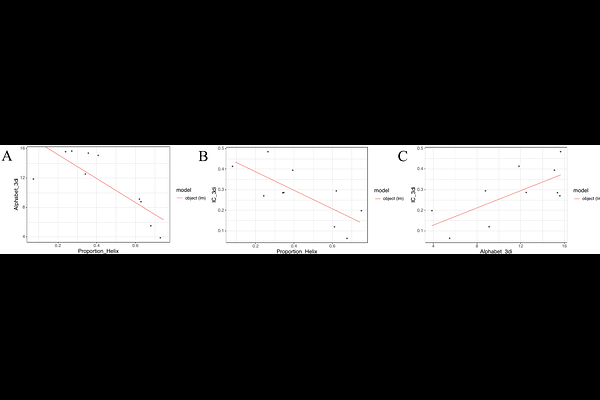
Adding 3Di characters to amino acid datasets can improve resolution, but the effect is weaker in shorter and alpha-helical proteins such as histones
Fullmer, M. S.; Puente-Lelievre, C.; Matzke, N. J.
AbstractThe recent introduction of Foldseek\'s 3Di character alphabet to encode 3D protein structure has opened up new possibilities for structural phylogenetics. The high conservation of structure relative to amino acid sequences raises the possibility of better resolving the deepest branches of life\'s evolution. As 3Di is a 20-letter alphabet of discrete characters, they are readily treatable with off-the-shelf algorithms for model-based phylogenetic inference and application of established methods such as bootstrapping. However, it remains to be seen if 3Di phylogenies are broadly more resolved than sequence-based phylogenies. We present data using samples from ten protein superfamilies showing that 3Di combines with sequence to produce better resolved phylogenies than either sequence or 3Di alone. We also show that information-theoretic measures, applied to superfamily alignments, significantly correlate with resolution in phylogenies derived from these alignments. Further, we identify the proportion of alpha helices in proteins as a major driver in reducing the information carried by 3Di character alignments, explaining the poorer performance of 3Di characters on some datasets with highly-conserved structure, such as Histone-fold proteins. Our results both provide encouragement for the further use of 3Di to address challenging questions in deep history, as well as sound a note of caution about which proteins it is suitable for.