Lizards on a sky archipelago: Genomic approaches to the evolution of the mountain genus Iberolacerta
Lizards on a sky archipelago: Genomic approaches to the evolution of the mountain genus Iberolacerta
Talavera, A.; Burriel-Carranza, B.; Mochales-Riano, G.; Estarellas, M.; Arribas, O.; Tejero-Cicuendez, H.; Salces-Ortiz, J.; Fernandez-Guiberteau, D.; Amat, F.; Niell, M.; Fernandez, R.; Mikheyev, A. S.; Carranza, S.
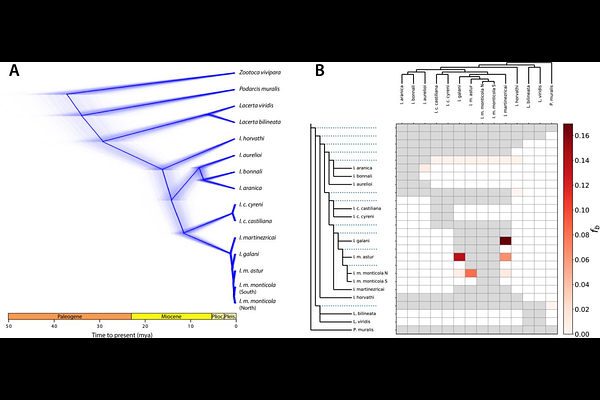
AbstractThe mountain-dwelling lizards of the genus Iberolacerta inhabit several isolated massifs across central and southwestern Europe. Their restricted and fragmented ranges, coupled with high altitude specialization in most species, entail a significant threat in the context of climate change for this group of lizards that has attracted interest from different fields. On the one hand, the alpine confinement of these relict species precedes the Pleistocene glacial cycles, and a few hypotheses have been proposed to explain it: from competitive exclusion by the wall lizards of the genus Podarcis, to adaptations to either cold or hypoxia, that would prevent them from expanding into lowlands. On the other hand, extensive research on chromosome evolution has shown Iberolacerta karyotypes to fairly differ from other lacertid lizards, exhibiting reductions in chromosome numbers and multiple sex chromosome determination systems. Here we present a chromosome-level genome assembly for Iberolacerta aurelioi, an Endangered rock lizard endemic to the Pyrenees. This genome has shed light on a genome architecture shaped by chromosome fusions, whose adaptive potential we discuss, as well as on expression shifts towards a hemoglobin isoform of enhanced oxygen affinity, as an adaptation to altitudinal hypoxia. In addition, medium coverage whole-genome sequencing data from 12 representatives encompassing all species and subspecies within the genus allowed us to address phylogenomic relationships, unveiling introgression events, gathering evidence in support of the competitive exclusion hypothesis through past demographic inference, and providing insights into homozygosity burdens, which offer valuable information for conservation efforts.