Identifying and quantifying the disease responses of different pennycress accessions to Alternaria japonica and Sclerotinia sclerotiorum
Identifying and quantifying the disease responses of different pennycress accessions to Alternaria japonica and Sclerotinia sclerotiorum
Kujur, A.; Codjoe, J. M.; Shah, D. M.; Chopra, R.
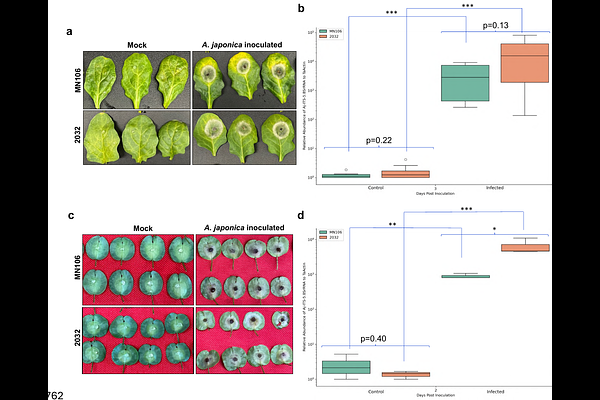
AbstractField Pennycress (Thlaspi arvense) is gaining attention in the US Midwest as a potential oilseed cover crop for their corn-soybean systems. Field research with breeding trials have shown that pennycress is susceptible to major fungal diseases affecting Brassica crops. In this study, we identified two pathogens: Alternaria japonica and Sclerotinia sclerotiorum which cause Alternaria black spot and Sclerotinia white mold diseases, respectively. Both fungi infect pennycress leaves and pods, with S. sclerotium also able to infect stems of the two pennycress accessions tested. We found accession 2032 to be more susceptible than MN106. Traditional visual methods to estimate disease severity could not capture the differential progression of Alternaria black spot in the two pennycress accessions. To address this, we developed a cost-effective DNA-based qPCR-based assay that can detect differential growth of both A. japonica and S. sclerotium on leaves and pods of two pennycress accessions. This method also provided more precise quantification of A. japonica and S. sclerotiorum at early infection stages than was possible visually. This assay could be helpful in evaluating various pennycress cultivars and other crops affected by these pathogens.