Characterization of Somatic Mutations in Human tRNA Genes Reveals Tumor-Specific Mutational Loads, and the Generation of tRNA Variants that Alter the Genetic Code
Characterization of Somatic Mutations in Human tRNA Genes Reveals Tumor-Specific Mutational Loads, and the Generation of tRNA Variants that Alter the Genetic Code
Murillo, M.; Salvadores, M.; Torres, A. G.; Vaquer Pico, A.; Tsapanou, L.; Supek, F.; Ribas de Pouplana, L.
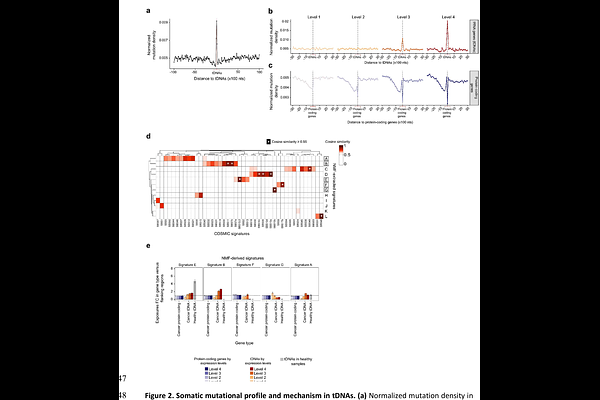
AbstractTransfer RNA genes (tDNAs) are essential genomic elements that safeguard translational fidelity. Using the T2T version of the human genome we have mapped the position of human tDNAs and analyzed their individual transcriptional activities. Then we have characterized, at single base resolution, the impact of somatic mutations in human tDNAs and its relationship to the transcriptional status of each gene. We confirm that tDNAs are hotspots for somatic mutagenesis, and show that they display mutational loads that are directly proportional to their transcription rates. Highly transcribed tDNAs in tumors or healthy tissues accumulate mutations at rates up to nine-fold higher than highly transcribed protein-coding genes. Mutational loads at tDNAs are tumor-specific, and increase with patient age. Mutations at structurally conserved tRNA positions appear to be under negative selection. Strikingly, anticodon nucleotides crucial for decoding frequently acquire somatic mutations, readily generating chimeric tRNAs species capable of systematically introducing amino acid substitutions across the proteome. Our results reveal a previously unrecognized source of somatic heterogeneity in human cancer and aging tissues that may directly impact upon translation efficiency and fidelity, and cause cell-specific proteostasis degeneration.