Codon-dependent regulation of mRNA translation and stability by ZC3H7A and ZC3H7B RNA-binding proteins
Codon-dependent regulation of mRNA translation and stability by ZC3H7A and ZC3H7B RNA-binding proteins
Harris Snell, P. L.; Naeli, P.; Garzia, A.; Waldron, J. A.; Chatterjee, S.; McGirr, T.; Ladak, R. J.; Choi, J.-H.; Luo, J.; Leino, S.; Jess, N.; Shariati, A.; Soto Rodriguez, X.; Gkogkas, C. G.; Sonenberg, N.; Tuschl, T.; Maguire, S.; Jafarnejad, S. M.
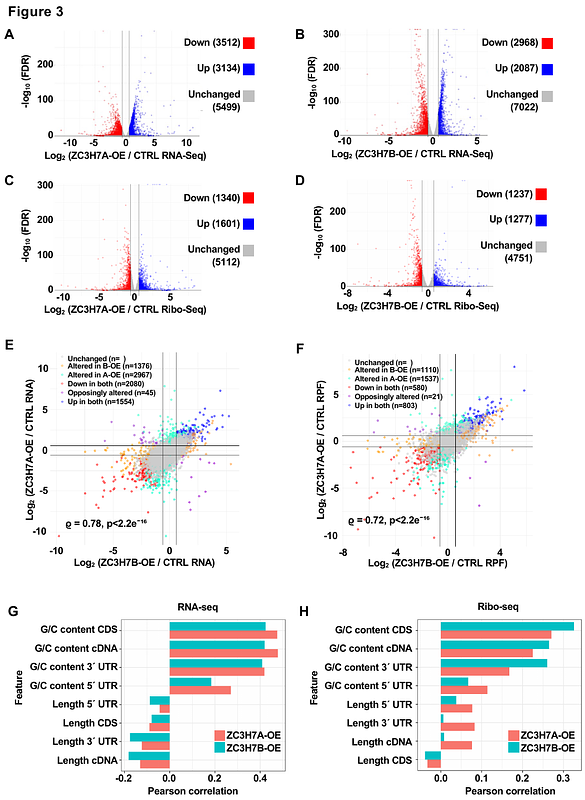
AbstractDecelerated translation elongation caused by non-optimal codons can reduce mRNA stability through codon optimality-mediated mRNA degradation. A key element of this process is the coupling of sensing the mRNA codon usage with the regulation of translation efficiency and stability. We report that two paralog RNA-binding proteins (ZC3H7A and ZC3H7B), which are only found in Chordates, preferentially bind to and reduce the stability and translation of mRNAs enriched in non-optimal codons with A/U at their wobble sites (A/U3 codons). ZC3H7A/B engage with ribosomes that lack elongation factors and induce mRNA degradation or block translation initiation through their interactions with the CCR4-NOT and the GIGYF2/4EHP translation repressor complex, respectively. Depletion of ZC3H7A and ZC3H7B or 4EHP impairs the repression of non-optimal A/U3-rich mRNAs. This study provides insights into a unique mechanism in higher eukaryotes that couples codon usage with the regulation of translation efficiency and mRNA stability.