Directing stem cell differentiation by chromatin state approximation
Directing stem cell differentiation by chromatin state approximation
Montano-Gutierrez, L. F.; Mueller, S.; Kutschat, A. P.; Seruggia, D.; Halbritter, F.
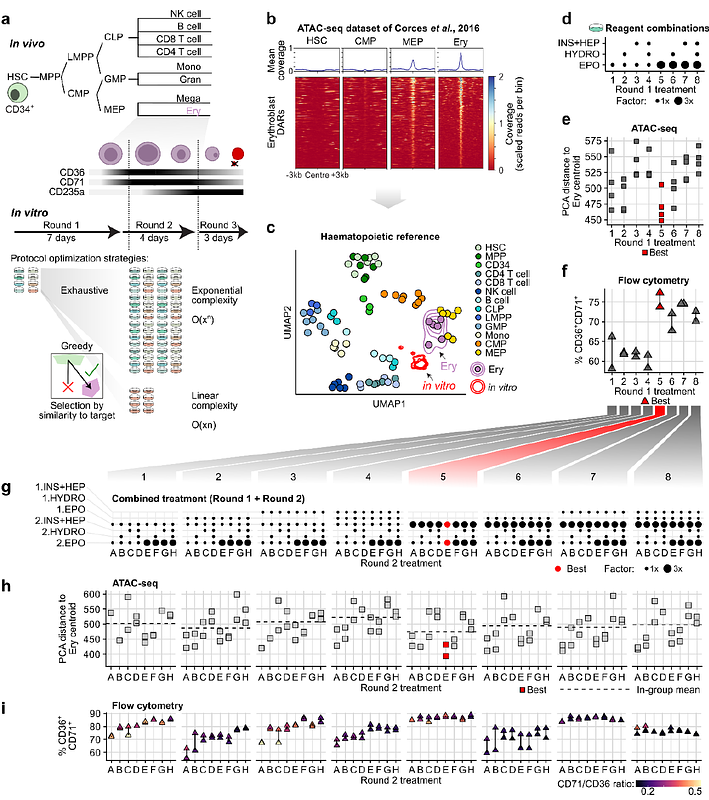
AbstractA prime goal of regenerative medicine is to replace dysfunctional cells in the body. To design protocols for producing target cells in the laboratory, one may need to consider exponentially large combinations of culture components. Here, we investigated the potential of iteratively approximating the target phenotype by quantifying the distance between chromatin profiles (ATAC-seq) of differentiating cells in vitro and their in-vivo counterparts. We tested this approach on the well-studied generation of erythroblasts from haematopoietic stem cells, evaluating a fixed number of components over two sequential differentiation rounds (8x8 protocols). We found that the most erythroblast-like cells upon the first round yielded the most erythroblast-like cells at the second round, suggesting that greedy selection by chromatin approximation can be a viable optimisation strategy. Furthermore, by analysing regulatory sequences in incompletely reprogrammed chromatin regions, we uncovered transcriptional regulators linked to roadblocks in differentiation and made a data-driven selection of ligands that further improved erythropoiesis. In future, our methodology can help craft notoriously difficult cells in vitro, such as B cells.