ProGuide: a flexible framework for modeling global conformational rearrangements in proteins using DEER-derived distance restraints
ProGuide: a flexible framework for modeling global conformational rearrangements in proteins using DEER-derived distance restraints
Grosskopf, J. D.; Kasson, P.; Lerch, M. T.
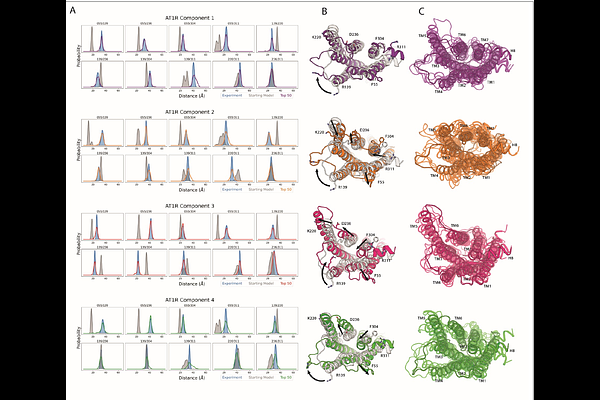
AbstractConformational heterogeneity is integral to protein function - ranging from enzyme catalysis to signal transduction - and visualizing distinct conformational states requires experimental techniques capable of providing such structural information. One particularly powerful method, double electron-electron resonance (DEER) spectroscopy, can provide a high-resolution, long-range (~15-80 [A]) probability distributions of distances between site-selected pairs of spin labels to resolve intra-protein distance parameters of unique protein conformations, as well as their respective likelihoods within a conformational ensemble. A current frontier in the field of DEER spectroscopy is utilizing this distance information in computational modeling to generate complete structural models of these multiple conformations. Although several methods have been developed for this purpose, modeling protein backbone structural rearrangements using multiple distance restraints remains challenging, due in part to the complexity provided by rotameric flexibility of the spin label side chain. Here, we overcome these challenges with ProGuide, a new framework for generating accurate structural models guided by DEER distance distribution information. Large conformational rearrangements are captured by performing iterative experimentally biased molecular dynamics simulations. In each iteration, spin-label rotameric heterogeneity is modeled using chiLife, and then C changes are calculated to capture distance-probability density present in the experimental DEER distributions and lacking from the modeled one. The resulting models of this process then go through a selection to generate the ensemble that best recapitulates the DEER data. We illustrate the power of this method using published DEER data from a study of biased agonism in the angiotensin II type 1 receptor (AT1R), a prototypical G protein coupled receptor (GPCR). The resulting AT1R models consist of both Gq- and {beta}-arrestin-biased conformations, including a completely novel {beta}-arrestin-biased conformation. These models reveal structural insights involving tertiary structural rearrangements as well as residue-level changes in crucial microswitch motifs. Taken together, the results demonstrate the power and flexibility of ProGuide to investigate conformational rearrangements of large, complex proteins using DEER-derived distance restraints.