Large Scale Cell Painting Guided Compound Selection Reveals Activity Cliffs and Functional Relationships
Large Scale Cell Painting Guided Compound Selection Reveals Activity Cliffs and Functional Relationships
Sanchez, M.; Bourriez, N.; Bendidi, I.; Cohen, E.; Svatko, I.; Del Nery, E.; Tajmouati, H.; Bollot, G.; Calzone, L.; Genovesio, A.
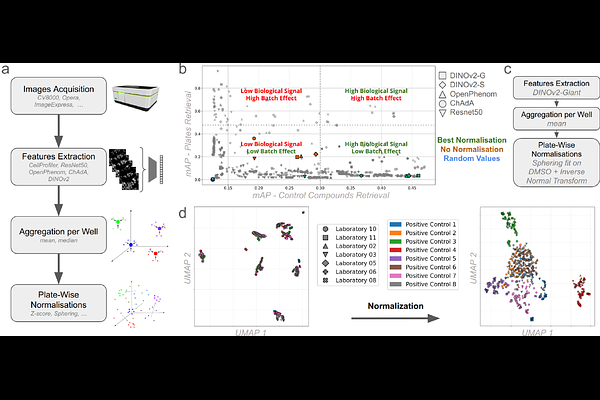
AbstractTraditional structure-based pre-screen compound selection relies on the assumption that chemical similarity implies similar biological activity. This paradigm narrows the exploration of chemical space and often fails to account for functional convergence, where structurally diverse compounds act through distinct targets to produce similar phenotypic effects. As a result, compounds with therapeutic potential may be overlooked. To overcome this constraint, we introduce a training-free, transfer learning-based method for large scale compound preselection that leverages deep phenotypic profiling of human cells. Notably, this enables robust pairwise comparison of phenotypic signatures across any source of the entire JUMP-CP, the largest publicly available cell painting dataset (112,480 compounds), preserving biological signals while mitigating batch effects. Validated across 65 high-throughput assays, including in vitro and in cellulo systems, our method provides efficient pre-screen enrichment of biologically active compounds, bypassing the blind spots of structure-centric approaches. Interestingly, because it is large scale, it also allows for a comprehensive analysis of structure-phenotypic activity relationships, revealing potentially thousands of compound activity cliffs, where minimal chemical changes in structure may result in profound phenotypic shifts. We show that these cliffs capture subtle, atom-level determinants of bioactivity that cannot be accessed by structure-based models. Furthermore, we demonstrate that structurally diverse compounds targeting different genes in the same biological pathway can induce either convergent or opposite phenotypes, a phenomenon validated across 30 pathways, hundreds of genes, and thousands of compounds. Finally, to support the broader community, we propose Phenoseeker, a web-based tool enabling instant retrieval of JUMP-CP compounds with similar phenotypic profiles. Together, these findings position phenotypic profiling not merely as a complementary tool, but as a transformative and scalable framework for navigating chemical space through a biological lens. By capturing rich morphological signatures that reflect functional outcomes, regardless of structural similarity, this approach enables the discovery of bioactive compounds, novel mechanisms of action, and unexpected target-pathway relationships. Applied at the scale of the JUMP-CP dataset, phenotypic profiling emerges as a powerful strategy for prioritizing compounds, illuminating activity cliffs, and accelerating the identification of therapeutically relevant candidates across diverse biological contexts.