Chromatin interaction-based annotation of distal cis-regulatory elements reveals highly dynamic promoter-enhancer interactions in lymphocyte development
Chromatin interaction-based annotation of distal cis-regulatory elements reveals highly dynamic promoter-enhancer interactions in lymphocyte development
Tingvall-Gustafsson, J.; Jensen, C. T.; Ungerback, J.; Sigvardsson, M.
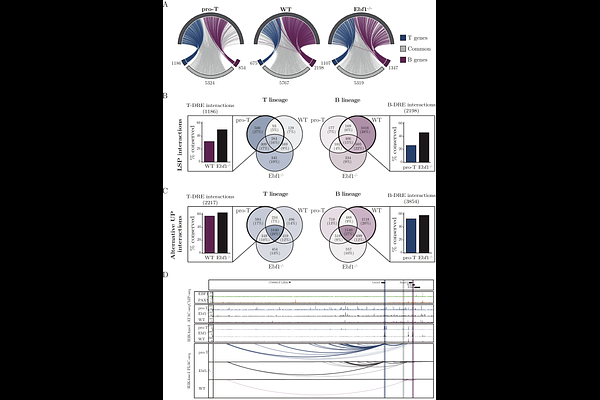
AbstractStage- and lineage-specific gene expression patterns are controlled by a complex interplay between transcription factors, the epigenetic landscape, and the 3-dimensional (3D) structure of the DNA. The 3D structure allows for the formation of DNA loops that juxtaposition distal regulatory elements to the promoters, allowing for tight control of gene expression. These loops can span hundreds of thousands base pairs, making it challenging to link regulatory elements to the correct target genes using conventional proximity-based annotation methods. Using a novel tool to facilitate the exploration of complex gene regulatory networks based on chromosome configuration data in early lymphocytes, we show that lineage-specific transcription factors target regulatory elements that are annotated to both lineage-specific and broadly expressed genes. Targeted inactivation of a set of these genes revealed their importance for B-cell development. Several regulatory elements annotated to lineage-specific genes were also annotated to alternative promoters in a context dependent manner, revealing a highly complex interplay between promoters and DREs in early lymphocyte development. Investigating the chromatin configuration in EBF1 deficient pro-B cells identified this transcription factor as a key mediator in the establishment of the 3-D promoter enhancer landscape in early B-lymphocyte differentiation. These data highlight how efficient annotation procedures for linking distal regulatory elements to target genes provide valuable insights into gene regulatory networks.