Spatial multi-omics and deep learning reveal fingerprints of immunotherapy response and resistance in hepatocellular carcinoma
Spatial multi-omics and deep learning reveal fingerprints of immunotherapy response and resistance in hepatocellular carcinoma
Wu, Z.; Boen, J.; Jindal, S.; Basu, S.; Bieniosek, M.; He, S.; LaPelusa, M. B.; Mayer, A. T.; Kaseb, A. O.; Zou, J.; Sharma, P.; Trevino, A. E.
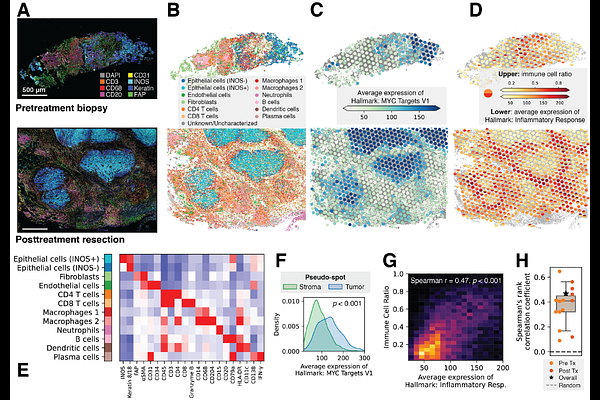
AbstractDespite advances in immunotherapy treatment, nonresponse rates remain high, and mechanisms of resistance to checkpoint inhibition remain unclear. To address this gap, we performed spatial transcriptomic and proteomic profiling on human hepatocellular carcinoma tissues collected before and after immunotherapy. We developed an interpretable, multimodal deep learning framework to extract key cellular and molecular signatures from these data. Our graph neural network approach based on spatial proteomic inputs achieved outstanding performance (ROC-AUC > 0.9) in predicting patient treatment response. Key predictive features and associated spatial transcriptomic profiles revealed the multi-omic landscape of immunotherapy response and resistance. One such feature was an interface niche expressing restrictive extracellular matrix factors that physically separates tumor tissue and lymphoid aggregates in nonresponders. We integrate this and other spatially-resolved signatures into SPARC, a multi-omic \"fingerprint\" comprising scores for immunotherapy response and resistance mechanisms. This study lays groundwork for future patient stratification and treatment strategies in cancer immunotherapy.