An Intrinsic-hoc Framework for Heterogeneous Cellular Senescence Elucidation Using Deep Graph Representation Learning and Experimental Validation
An Intrinsic-hoc Framework for Heterogeneous Cellular Senescence Elucidation Using Deep Graph Representation Learning and Experimental Validation
Ma, A.; Cheng, H.; Vanegas, N. D. P.; Ghobashi, A.; Chen, H.; Rodriguez, J.; Rosas, L.; Wang, C.; Shao, J.; Jiang, Y.; Wang, X.; Rahman, I.; Lugo-Martinez, J.; Li, D.; Pryhuber, G. S.; Bar-Joseph, Z.; Eickelberg, O.; Konigshoff, M.; Chung, D.; Chen, R.; Finkel, T.; Mora, A.; Rojas, M.; Ma, Q.
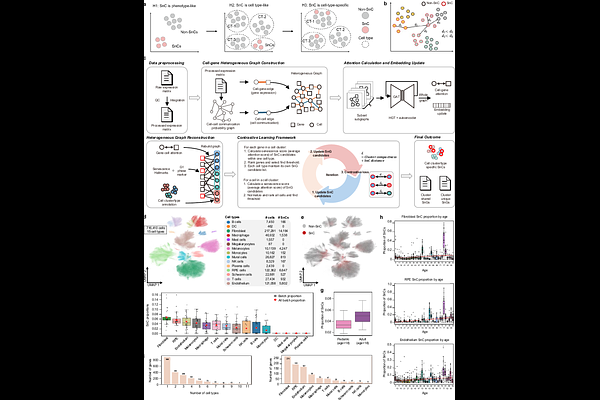
AbstractCharacterizing senescent cells and identifying corresponding senescence-associated genes within complex tissues is critical for our understanding of aging and age-related diseases. We present DeepSAS, an intrinsic-hoc framework for elucidating the heterogeneity encoded in senescent cells and their associated genes from single-cell RNA-seq data using deep graph representation learning. Applied to both healthy eye cell atlas and in-house idiopathic pulmonary fibrosis datasets with Xenium spatial transcriptomics validation, DeepSAS reveals robust and biologically grounded senotypes and demonstrates superior benchmarking performance compared with existing methods.